细菌通常分布于皮肤表面、呼吸系统,以及外分泌器官如乳腺导管、阴道和胃肠道等[1]。据估计,全世界约20%的肿瘤由细菌引起[2]。细菌可穿过胃肠黏膜而暴露于肠肝循环[3],少量细菌可能会出现在血液循环中,甚至有证据表明,由于异常的肿瘤脉管系统的居留和外渗作用,一些细菌可聚集在肿瘤内。基于二代测序技术的细菌16S RNA测序可鉴定许多通常条件下不可培养的细菌,从而大大提升了对生物体微环境变化引起的代谢与菌群响应机制的认识[4]。随着高通量测序技术的发展,涌现出了更好的序列拼接技术、更完整的数据库[5]及更深度的测序方法[6],进一步推动了直至单个细菌的识别和量化[7]。利用宏基因组技术研究发现,与癌旁组织或患者健康组织相比,多种癌症中发现了新的含量丰富的病原菌。另外,传统上被认为是无菌的肿瘤生长部位也有细菌DNA指征。近年来已在多处鉴定出肿瘤特异的细菌群落,如结肠[8]、喉部[9]、胰腺[10]和前列腺[11]等。然而,在上述关联研究中,一个主要的问题是,这些细菌是作为一个旁观者生存于肿瘤生态位还是会对肿瘤的发病或肿瘤的持续性有影响。本文就肠道细菌与癌症发生发展及治疗之间的关系进行综述,同时关注少量肿瘤的自身细菌。
1 人体肠道细菌概述正常情况下,成年人肠道细菌数量为1013~1014个,与人体细胞总数相近[12]。人肠道菌群中有4种占主导地位,其中厚壁菌门(Firmicutes)和拟杆菌门(Bacteroidetes)约占90%,其次为变形菌门(Proteobacteria)和放线菌门(Actinomycetes),含量较低[13]。厚壁菌主要为革兰阳性菌,包括厌氧梭状芽胞杆菌(Clostridium)、链球菌(Streptococcus)和肠球菌(Enterococcus)。拟杆菌是一种可代谢复合糖类(碳水化合物)的革兰阴性菌,包括已知的多形拟杆菌(Bacteroides thetaiotamicron)和脆弱拟杆菌(Bacteroides fragilis)。变形菌是一组革兰阴性菌,包括γ变形菌(Gammaproteobacteria)、大肠埃希菌(Escherichia coli)和克雷伯菌(Klebsiella species)。通常个体细菌组成比较稳定,偶尔会因抗生素使用而受到较大干扰[4]。大部分细菌可在上皮黏膜层的保护下正常繁殖[13],只有少数易变化的菌种可能对维持个体细菌种群的完整和健康至关重要[14]。
随着对肠道细菌组分的逐步了解,人们发现细菌能与抗原呈递细胞直接相互作用及通过代谢物(尤其是短链脂肪酸,如丁酸)相互作用,对宿主上皮表面的天然免疫和适应性免疫系统进行信号转导调节[15]。此外,一些细菌可合成直接作用于人类受体的化合物[16]。反之,宿主也可通过抗菌肽[17]、灭活IgA[18]和代谢物(如黏蛋白类聚糖)等,优先保留或排斥某些细菌[19]。
2 肠道细菌与癌症的相关性 2.1 肠道细菌与结直肠癌 2.1.1 细菌与结直肠癌的发生结直肠是哺乳动物体内细菌群落密度最高的场所。多项研究显示,与健康或癌前炎症的结直肠组织相比,结直肠癌样本中[8, 20]梭杆菌(Fusobacterium species)明显增多,且不受地理位置、患者年龄、测序技术等影响。
对梭杆菌所在肿瘤微环境的研究表明,其可上调骨髓来源抑制性细胞(myeloid-derived suppressor cell,MDSC)。MDSC已被证明可促进肿瘤形成并恶化预后,其部分机制是通过干扰免疫监视,即逃离免疫系统的监视而促进肿瘤发生[21]。此外,体外研究表明梭杆菌能通过Fap2黏附蛋白与含Ig和ITIM结构域的T细胞免疫受体(T-cell immunoreceptor with Ig and ITIM domains,TIGIT)结合[22]。TIGIT是自然杀伤(natural killer,NK)细胞的标记蛋白,其激活会增强对肿瘤的杀伤,而梭杆菌与TIGIT的结合导致NK细胞的抗肿瘤活性被阻断,从而帮助肿瘤逃避免疫破坏[23]。为进一步逃避免疫,梭杆菌还可存活于活细胞内,通过其表面蛋白FadA与宿主上皮细胞E-钙黏蛋白结合,促进细胞摄取和梭杆菌侵入。随后,这种相互作用诱导结肠癌细胞株HCT116产生炎性细胞因子如白细胞介素6(interleukin 6,IL-6)、IL-8和IL-18,而阻断FadA介导的细胞对梭杆菌的摄取会阻止细胞因子产生,且缺乏FadA的非侵袭性梭杆菌也不会触发炎症反应[24],表明该炎症效应完全由梭杆菌侵入宿主上皮细胞引起。
梭杆菌也可独立于免疫系统而促进肿瘤发生。例如,梭杆菌与E-钙黏蛋白结合诱导促β-catenin信号转导,导致肿瘤发生[24]。分别使用梭杆菌阳性和阴性肿瘤建立异种移植小鼠模型时,发现只有梭杆菌阳性肿瘤能移植成功,且经过4代繁殖的肿瘤仍保留部分梭杆菌。以甲硝唑治疗后,梭杆菌数量显著下降,同时伴随肿瘤增殖能力下降。这表明梭杆菌在免疫系统中以免疫非依赖性方式促进肿瘤生长,且生物体与梭杆菌相关癌症之间存在共生关系[25]。梭杆菌与人体内炎性微环境亦相关,即肿瘤组织中梭杆菌增加会提高核因子κB(nuclear factor κB,NF-κB)转录水平,同时降低CD3+ T细胞数量,导致预后更差[20, 24],进一步表明梭杆菌与缺乏抗肿瘤免疫细胞机制的肿瘤发生相关[26]。此外,梭杆菌水平升高伴随拟杆菌和双歧杆菌(Bifidobacteriaceae)降低的克罗恩病患者,具有更高的胃肠道及其他常见癌症的发病率[27]。
2.1.2 细菌代谢物与结直肠癌的发生多种细菌已被证实能分泌细菌毒素及其他致癌物。例如,带有pks基因簇的肠毒性大肠埃希菌会产生一种聚酮糖,它能诱导哺乳动物细胞DNA双链断裂[28]。用含有pks的大肠埃希菌饲喂易患结直肠癌的IL-10基因敲除小鼠,可导致肿瘤发生;而使用其他致炎性细菌如粪肠球菌或缺乏pks的大肠埃希菌,可引起类似的炎症状况,但不引发肿瘤形成[29]。有研究显示,21例结直肠癌患者中有14例黏膜相关结肠组织中含有毒性pks基因簇,而在24例健康对照者中仅有5例具有该基因簇,进一步证实pks与结直肠癌具有高度相关性[29]。与此相反,在APCmin/+模型小鼠中,脆弱拟杆菌产生的肠毒素通过Stat3活化激活Th17表型这种炎症依赖机制来诱导肿瘤发生[30]。因此,细菌可能会通过特定的细菌毒素来增加癌变风险,或者可能通过促炎作用加速这一过程。
再如,具核梭杆菌(Fusobacterium nucleatum)的分泌物在口服给药时可加剧APCmin/+模型小鼠的肿瘤恶化,但不加剧仅有炎症的IL-10缺失小鼠或Tbet/Rag2缺失小鼠的肿瘤发生[25]。这表明具核梭杆菌在肿瘤易发模型中促进炎症并加速肿瘤发生,但在这些模型中炎症恶化不是致癌性的[20]。
此外,细菌与结肠癌关联性研究显示,采用植物性饮食的人具有较低的结肠癌发病率[31]。原因可能是以植物为基础的饮食可使细菌合成更多的短链脂肪酸如醋酸盐、丙酸盐和丁酸盐,这些脂肪酸可诱导大肠组织产生调节性T细胞而发挥抗炎作用。
2.1.3 细菌与结直肠癌的治疗与APC蛋白功能丧失致癌相似,在Cdx2诱导的APC小鼠结肠癌模型中,小鼠远端结肠发生癌变,且肿瘤细胞显现炎性IL-23/IL-17特征,而抗生素治疗与IL-23受体敲除均能有效减小肿瘤体积,该过程可能与Toll样受体2(Toll-like receptor 2,TLR2)和髓样分化因子初级应答基因88(myeloid differentiation primary response 88,MYD88)信号转导的天然免疫激活相关[32]。再以伊立替康(irinotecan)为例,在一些患者中细菌合成的β-葡萄糖醛酸酶可在结肠中重新激活药物而导致更严重的药物不良反应,如剂量限制性腹泻等[33]。此外,环磷酰胺也依赖细菌免疫机制发挥作用,其通过直接损伤小肠上皮诱导肠道泄漏,从而使肠道细菌向肠道相关淋巴组织转运并激活抗肿瘤Th17应答[34]。
2.2 肠道细菌与黏膜相关淋巴组织(mucosa-associated lymphoid tissue,MALT)淋巴瘤和胃腺癌 2.2.1 细菌通过多种机制诱发淋巴癌和胃癌幽门螺杆菌(Helicobacter pylori)作为Ⅰ类致癌物[2],能入侵并定植于宿主,控制免疫系统及驱动癌症关键标记基因,如促进炎-癌反应、破坏抗肿瘤免疫和上调细胞增殖信号,进而导致胃MALT淋巴瘤和胃腺癌发生[35]。
幽门螺杆菌可持久性诱发肿瘤的形成。其能借助鞭毛穿过胃黏膜层以及多种黏附素(如SabA和BabA[36-37])黏附于胃上皮细胞。一旦幽门螺杆菌与上皮紧密相连,其会利用Ⅳ型分泌系统将CagA和其他毒力因子沉积于细胞。CagA进入细胞后对SHP2(一种宿主癌蛋白)进行调节,导致细胞不受控制地生长和运动[38]。此外,其他毒力因子如VacA可在宿主细胞膜上形成孔道,诱导细胞死亡和更高的细胞自我更新频率[39]。
除产毒因子直接影响上皮细胞外,幽门螺杆菌还能诱导天然免疫炎症信号导致慢性炎症。幽门螺杆菌感染可诱导中性粒细胞和巨噬细胞产生活性氧(reactive oxygen species,ROS)及炎性细胞因子,如IL-1β、肿瘤坏死因子α(tumor necrosis factor α,TNF-α)和IL-8,最终导致肠道内Th1免疫应答。在此过程中,与其他病原体类似,幽门螺杆菌通过自身过氧化氢酶和超氧化物歧化酶保护自身免受ROS影响,但使宿主组织细胞DNA受到损伤,最终导致宿主基因发生突变。此外,幽门螺杆菌感染可提高NF-κB表达水平,进一步推动与肿瘤发生发展相关的炎症反应[40]。
除诱导天然免疫外,幽门螺杆菌还会针对T细胞产生适应性免疫。例如,毒力因子VacA可阻断活化T细胞核因子(nuclear factor of activated T cells,NFAT)激活,进而使IL-2诱导的T细胞增殖受损,导致幽门螺杆菌被清除[39]。此外,幽门螺杆菌产生的γ-谷氨酰转移酶也能阻断T细胞增殖[41],并激活Th1和Th17免疫应答,这对产生持续性感染非常重要,且可能降低对肿瘤的免疫监视。幽门螺杆菌还可诱导程序性死亡配体1(programmed death ligand 1,PD-L1)在胃上皮细胞的表达,进而调控T细胞程序性死亡受体1(programmed cell death 1,PD-1),导致免疫监视丧失[42]。因此,细胞损伤、固有致癌信号产生和免疫监视降低都会促使部分患者发生癌症。
2.2.2 细菌与淋巴癌和胃癌的治疗某些癌症可能会严重依赖其微环境中的细菌而持续生长并逃避免疫系统,因为患者接受抗生素治疗后,肿瘤可得到有效控制。最显著的例子是使用兰索拉唑30 mg、阿莫西林1 g和克拉霉素500 mg(简称PREVPAC)联合治疗幽门螺杆菌诱导的MALT淋巴瘤[43],以及使用质子泵抑制剂、阿莫西林和克拉霉素联合治疗幽门螺杆菌引起的胃癌[44]。此外,鹦鹉热衣原体可引发眼附件MALT淋巴瘤。在44例患者中有39例检测出衣原体感染,34例采用多西环素治疗后,6例肿瘤完全缓解,16例肿瘤部分缓解。这也如预期所料,即衣原体减少和去除状态与肿瘤消退程度相一致[45]。
2.3 肠道细菌与其他癌症 2.3.1 前列腺癌与细菌相关的肿瘤发生并不局限于胃肠道。Golombos等研究发现,胰腺癌患者的粪便中Bacteroides massiliensis显著增高[10]。此外,有报道证明痤疮丙酸杆菌(Propionibacterium acnes)参与前列腺癌的发生[46]。有趣的是,这些痤疮丙酸杆菌与生长在皮肤表面的痤疮丙酸杆菌具有完全不同的特性,它们能感染、入侵宿主细胞并诱导环氧合酶2(cyclooxygenase 2,COX-2)信号转导而促进细胞增殖。将前列腺癌患者的痤疮丙酸杆菌植入小鼠前列腺后,可诱发炎症及加速细胞增殖[47]。
2.3.2 肝癌越来越多的证据表明,肠道细菌能促进肝癌的发展。肠道细菌生态失调、代谢物改变等均会加重肝脏炎症、纤维化或基因毒性,从而促进肝癌形成[48]。另外,细菌可通过影响宿主胆汁酸代谢促进肝癌发生。Yoshimoto等[49]和Quante等[50]报道,梭杆菌可将人体次生胆汁酸转化为脱氧胆酸,脱氧胆酸具有高致癌性,可诱导肝星状细胞衰老和肝细胞DNA损伤。高脂肪饮食可导致Ⅺ和Ⅹ Ⅳ亚群的梭杆菌富集而改变胆汁代谢,产生更多脱氧胆酸,最终促使非酒精性脂肪肝发展为肝癌。使用梭杆菌敏感的抗生素如万古霉素治疗,可预防肿瘤发生;同时,在有抗生素的情况下,额外添加脱氧胆酸可恢复肿瘤形成,进一步表明梭杆菌产生的脱氧胆酸是致癌物。此外,在单剂量二乙基亚硝胺和重复剂量四氯化碳处理炎性纤维化肝癌小鼠模型后,使用抗生素部分去除肠道细菌可抑制癌变,即使纤维化已很明显,抗生素治疗仍有效。由于肝癌的发生依赖肝细胞TLR4激活,敲除TLR4也会减少小鼠肿瘤数目及缩小体积。TLR4的主要配体是脂多糖,表明细菌来源的脂多糖可增强肿瘤发生[51]。
2.3.3 胰腺癌研究表明,肠道细菌如幽门螺杆菌感染会增加胰腺癌的发病风险。幽门螺杆菌感染可致胃酸分泌减少,不仅会提高亚硝基类化合物的合成,同时胃酸不足还会使胃腔清理不彻底而改变肠道细菌组成,两者共同作用提高了胰腺癌的发病率[52]。近期,Golombos等进一步证明肠道细菌可迁移到胰腺中,且胰腺癌小鼠模型和胰腺癌患者胰腺中的细菌丰度均高于相应对照组。与正常小鼠相比,胰腺癌模型小鼠中肠道细菌迁移至胰腺的能力增强。另外,在无菌突变小鼠中,胰腺癌的发病较为缓慢,且口服抗生素可进一步延缓肿瘤生长,而饲料中添加移植胰腺癌小鼠的粪便又会加速肿瘤进展。此外,该实验还发现了一些特定的细菌如变形菌、放线菌、梭杆菌和疣微菌(Verrucomicrobia),它们在胰腺癌患者的肠道中显著增多,变形菌在胰腺中也大量富集,而假长双歧杆菌(Bifidobacterium pseudolongum)则能加速致癌作用[10]。
肠道细菌亦可干预肿瘤化疗效果。最新研究表明,人胰腺癌组织细胞中含有支原体和γ变形菌,将这些细菌移植到小鼠体内会诱导产生对化疗药物吉西他滨的耐药性,而用环丙沙星进行抗菌治疗后,抗肿瘤效应得以解除[53]。
2.3.4 黑色素瘤黑色素瘤是来源于黑色素细胞的一类恶性肿瘤,是皮肤肿瘤中恶性程度最高的瘤种,其被发现时往往已发生肿瘤转移。使用免疫检查点抑制剂靶向细胞毒性T细胞相关蛋白4(cytotoxic T-lymphocyte-associated protein 4,CTLA4)治疗可明显延长患者存活时间,但近一半患者对免疫检查点抑制剂不敏感。
为探讨具体原因,Vétizou等[54]和Sivan等[55]通过小鼠模型先后发现了决定小鼠同种移植肿瘤对CTLA4和PD-L1抑制干扰的细菌。Vétizou等对不同小鼠同种异体移植模型的研究表明,大部分CTLA4抑制效果可被抗生素消除,但可通过特定的细菌,特别是脆弱拟杆菌来恢复。用脆弱拟杆菌口服或用脆弱拟杆菌来源的多糖进行免疫治疗都可发挥抗肿瘤作用。Sivan等发现,两组来自不同供应商的C57/B16小鼠中,PD-L1抑制剂只对一组的B16黑色素瘤生长有抑制作用。而将来自两个供应商的小鼠共同饲养时,两组均表现出对PD-L1抑制剂的反应。进一步研究发现,双歧杆菌参与了小鼠共居转移,且口服该菌有效,但热灭活后口服无效。此外,抗肿瘤活性结果表明,小鼠体内存在一种可交叉呈递抗原的树突细胞,其能增强已激活的CD8+ T细胞的浸润性。
另外,两项最新有关黑色素瘤的研究揭示,不同生物体与不同细菌组合也会造成对PD-1抑制剂反应(反应与非反应)的差异[56-57]。其中一项研究表明,多形拟杆菌与非反应相关,但与小鼠中多形拟杆菌具有保护作用的结论相反。总之,肠道细菌介导的免疫性反应可严重影响免疫检查点抑制剂的作用效果,利用特定细菌调节该过程可能是优化检查点抑制剂反应的有效方式。
3 结语宿主细菌与癌症之间的相互作用多种多样,细菌通过各种途径影响癌症进展,包括直接作用、通过代谢物(毒素)及天然和适应性免疫作用(图 1)。至今,仅有少数细菌及其致癌或抗癌机制被阐明。随着对人体细菌分类和代谢分布的深入了解,有望实现解析特定细菌与癌症发生或抗癌干预的相关性。解析肠道细菌与癌症详细作用机制的良好开端,预示着从肠道细菌发现新的癌症诊断和治疗靶点的巨大潜能。
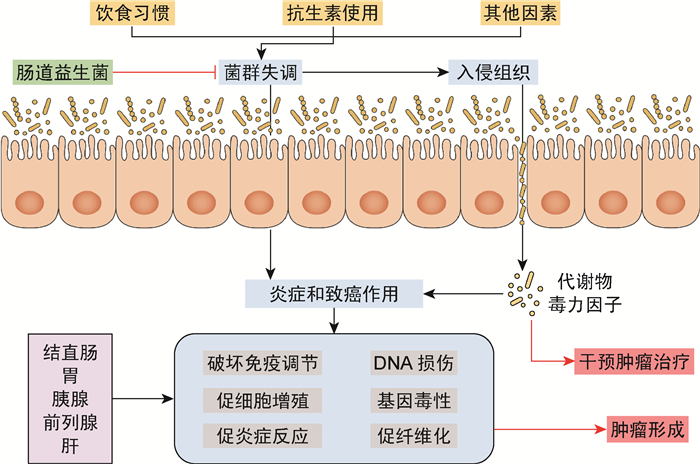
|
| 图 1 肠道细菌与癌症的关系 Fig. 1 The relationship between gut microbiota and cancer |
| [1] |
Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, Mohan N, Aykut B, Usyk M, Torres LE, Werba G, Zhang K, Guo Y, Li Q, Akkad N, Lall S, Wadowski B, Gutierrez J, Kochen Rossi JA, Herzog JW, Diskin B, Torres-Hernandez A, Leinwand J, Wang W, Taunk PS, Savadkar S, Janal M, Saxena A, Li X, Cohen D, Sartor RB, Saxena D, Miller G. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression[J]. Cancer Discov, 2018, 8(4): 403-416.
[DOI]
|
| [2] |
de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis[J]. Lancet Oncol, 2012, 13(6): 607-615.
[DOI]
|
| [3] |
Clavel T, Gomes-Neto JC, Lagkouvardos I, Ramer-Tait AE. Deciphering interactions between the gut microbiota and the immune system via microbial cultivation and minimal microbiomes[J]. Immunol Rev, 2017, 279(1): 8-22.
[DOI]
|
| [4] |
Raymond F, Ouameur AA, Déraspe M, Iqbal N, Gingras H, Dridi B, Leprohon P, Plante PL, Giroux R, Bérubé È, Frenette J, Boudreau DK, Simard JL, Chabot I, Domingo MC, Trottier S, Boissinot M, Huletsky A, Roy PH, Ouellette M, Bergeron MG, Corbeil J. The initial state of the human gut microbiome determines its reshaping by antibiotics[J]. ISME J, 2016, 10(3): 707-720.
[DOI]
|
| [5] |
Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments[J]. Genome Biol, 2014, 15(3): R46.
[DOI]
|
| [6] |
Wooley JC, Ye Y. Metagenomics: facts and artifacts, and computational challenges[J]. J Comput Sci Technol, 2010, 25(1): 71-81.
[DOI]
|
| [7] |
Scarpellini E, Ianiro G, Attili F, Bassanelli C, De Santis A, Gasbarrini A. The human gut microbiota and virome: potential therapeutic implications[J]. Dig Liver Dis, 2015, 47(12): 1007-1012.
[DOI]
|
| [8] |
Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, Tjalsma H. Towards the human colorectal cancer microbiome[J]. PLoS One, 2011, 6(5): e20447.
[DOI]
|
| [9] |
Gong HL, Shi Y, Zhou L, Wu CP, Cao PY, Tao L, Xu C, Hou DS, Wang YZ. The composition of microbiome in larynx and the throat biodiversity between laryngeal squamous cell carcinoma patients and control population[J]. PLoS One, 2013, 8(6): e66476.
[DOI]
|
| [10] |
Golombos DM, Ayangbesan A, O'Malley P, Lewicki P, Barlow L, Barbieri CE, Chan C, DuLong C, Abu-Ali G, Huttenhower C, Scherr DS. The role of gut microbiome in the pathogenesis of prostate cancer: a prospective, pilot study[J]. Urology, 2018, 111: 122-128.
[DOI]
|
| [11] |
Doorakkers E, Lagergren J, Engstrand L, Brusselaers N. Helicobacter pylori eradication treatment and the risk of gastric adenocarcinoma in a Western population[J]. Gut, 2018.
[DOI]
|
| [12] |
Pabst O. Correlation, consequence, and functionality in microbiome-immune interplay[J]. Immunol Rev, 2017, 279(1): 4-7.
[DOI]
|
| [13] |
Kim S, Covington A, Pamer EG. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens[J]. Immunol Rev, 2017, 279(1): 90-105.
[DOI]
|
| [14] |
Claussen JC, Skiecevičienė J, Wang J, Rausch P, Karlsen TH, Lieb W, Baines JF, Franke A, Hütt MT. Boolean analysis reveals systematic interactions among low-abundance species in the human gut microbiome[J]. PLoS Comput Biol, 2017, 13(6): e1005361.
[DOI]
|
| [15] |
Ilinskaya ON, Ulyanova VV, Yarullina DR, Gataullin IG. Secretome of intestinal bacilli: a natural guard against pathologies[J]. Front Microbiol, 2017, 8: 1666.
[DOI]
|
| [16] |
Cohen LJ, Esterhazy D, Kim SH, Lemetre C, Aguilar RR, Gordon EA, Pickard AJ, Cross JR, Emiliano AB, Han SM, Chu J, Vila-Farres X, Kaplitt J, Rogoz A, Calle PY, Hunter C, Bitok JK, Brady SF. Commensal bacteria make GPCR ligands that mimic human signalling molecules[J]. Nature, 2017, 549(7670): 48-53.
[DOI]
|
| [17] |
Britanova L, Diefenbach A. Interplay of innate lymphoid cells and the microbiota[J]. Immunol Rev, 2017, 279(1): 36-51.
[DOI]
|
| [18] |
McCoy KD, Ronchi F, Geuking MB. Host-microbiota interactions and adaptive immunity[J]. Immunol Rev, 2017, 279(1): 63-69.
[DOI]
|
| [19] |
Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease[J]. Immunol Rev, 2017, 279(1): 70-89.
[DOI]
|
| [20] |
Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment[J]. Cell Host Microbe, 2013, 14(2): 207-215.
[DOI]
|
| [21] |
Montero AJ, Diaz-Montero CM, Kyriakopoulos CE, Bronte V, Mandruzzato S. Myeloid-derived suppressor cells in cancer patients: a clinical perspective[J]. J Immunother, 2012, 35(2): 107-115.
[DOI]
|
| [22] |
Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklić K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack[J]. Immunity, 2015, 42(2): 344-355.
[DOI]
|
| [23] |
Blake SJ, Dougall WC, Miles JJ, Teng MW, Smyth MJ. Molecular pathways: targeting CD96 and TIGIT for cancer immunotherapy[J]. Clin Cancer Res, 2016, 22(21): 5183-5188.
[DOI]
|
| [24] |
Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin[J]. Cell Host Microbe, 2013, 14(2): 195-206.
[DOI]
|
| [25] |
Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, Chipashvili O, Hagan T, Walker M, Ramachandran A, Diosdado B, Serna G, Mulet N, Landolfi S, Ramon Y, Cajal S, Fasani R, Aguirre AJ, Ng K, Élez E, Ogino S, Tabernero J, Fuchs CS, Hahn WC, Nuciforo P, Meyerson M. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer[J]. Science, 2017, 358(6369): 1443-1448.
[DOI]
|
| [26] |
Mima K, Cao Y, Chan AT, Qian ZR, Nowak JA, Masugi Y, Shi Y, Song M, da Silva A, Gu M, Li W, Hamada T, Kosumi K, Hanyuda A, Liu L, Kostic AD, Giannakis M, Bullman S, Brennan CA, Milner DA, Baba H, Garraway LA, Meyerhardt JA, Garrett WS, Huttenhower C, Meyerson M, Giovannucci EL, Fuchs CS, Nishihara R, Ogino S. Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location[J]. Clin Transl Gastroenterol, 2016, 7(11): e200.
[DOI]
|
| [27] |
Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, González A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier R, Xavier RJ. The treatment-naive microbiome in new-onset Crohn's disease[J]. Cell Host Microbe, 2014, 15(3): 382-392.
[DOI]
|
| [28] |
Nougayrède JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells[J]. Science, 2006, 313(5788): 848-851.
[DOI]
|
| [29] |
Arthur JC, Gharaibeh RZ, Mühlbauer M, Perez-Chanona E, Uronis JM, McCafferty J, Fodor AA, Jobin C. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer[J]. Nat Commun, 2014, 5: 4724.
[DOI]
|
| [30] |
Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, Housseau F, Pardoll DM, Sears CL. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses[J]. Nat Med, 2009, 15(9): 1016-1022.
[DOI]
|
| [31] |
Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis[J]. Science, 2013, 341(6145): 569-573.
[DOI]
|
| [32] |
Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung KE, Datz C, Feng Y, Fearon ER, Oukka M, Tessarollo L, Coppola V, Yarovinsky F, Cheroutre H, Eckmann L, Trinchieri G, Karin M. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth[J]. Nature, 2012, 491(7423): 254-258.
[DOI]
|
| [33] |
Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, Venkatesh M, Jobin C, Yeh LA, Mani S, Redinbo MR. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme[J]. Science, 2010, 330(6005): 831-835.
[DOI]
|
| [34] |
Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ, Schlitzer A, Ginhoux F, Apetoh L, Chachaty E, Woerther PL, Eberl G, Bérard M, Ecobichon C, Clermont D, Bizet C, Gaboriau-Routhiau V, Cerf-Bensussan N, Opolon P, Yessaad N, Vivier E, Ryffel B, Elson CO, Doré J, Kroemer G, Lepage P, Boneca IG, Ghiringhelli F, Zitvogel L. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide[J]. Science, 2013, 342(6161): 971-976.
[DOI]
|
| [35] |
Peek RM Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas[J]. Nat Rev Cancer, 2002, 2(1): 28-37.
[DOI]
|
| [36] |
Styer CM, Hansen LM, Cooke CL, Gundersen AM, Choi SS, Berg DE, Benghezal M, Marshall BJ, Peek RM Jr, Borén T, Solnick JV. Expression of the BabA adhesin during experimental infection with Helicobacter pylori[J]. Infect Immun, 2010, 78(4): 1593-1600.
[DOI]
|
| [37] |
Mahdavi J, Sondén B, Hurtig M, Olfat FO, Forsberg L, Roche N, Angstrom J, Larsson T, Teneberg S, Karlsson KA, Altraja S, Wadström T, Kersulyte D, Berg DE, Dubois A, Petersson C, Magnusson KE, Norberg T, Lindh F, Lundskog BB, Arnqvist A, Hammarström L, Borén T. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation[J]. Science, 2002, 297(5581): 573-578.
[DOI]
|
| [38] |
Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein[J]. Nat Rev Cancer, 2004, 4(9): 688-694.
[DOI]
|
| [39] |
Sundrud MS, Torres VJ, Unutmaz D, Cover TL. Inhibition of primary human T cell proliferation by Helicobacter pylori vacuolating toxin (VacA) is independent of VacA effects on IL-2 secretion[J]. Proc Natl Acad Sci USA, 2004, 101(20): 7727-7732.
[DOI]
|
| [40] |
Salama NR, Hartung ML, Müller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori[J]. Nat Rev Microbiol, 2013, 11(6): 385-399.
[DOI]
|
| [41] |
Schmees C, Prinz C, Treptau T, Rad R, Hengst L, Voland P, Bauer S, Brenner L, Schmid RM, Gerhard M. Inhibition of T-cell proliferation by Helicobacter pylori γ-glutamyl transpeptidase[J]. Gastroenterology, 2007, 132(5): 1820-1833.
[DOI]
|
| [42] |
Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy[J]. Nat Rev Cancer, 2012, 12(4): 252-264.
[DOI]
|
| [43] |
Stolte M, Bayerdörffer E, Morgner A, Alpen B, Wündisch T, Thiede C, Neubauer A. Helicobacter and gastric MALT lymphoma[J]. Gut, 2002, 50(Suppl 3): III19-III24.
[PubMed]
|
| [44] |
Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation[J]. Nat Rev Gastroenterol Hepatol, 2017, 14(9): 527-539.
[DOI]
|
| [45] |
Ferreri AJ, Govi S, Pasini E, Mappa S, Bertoni F, Zaja F, Montalbán C, Stelitano C, Cabrera ME, Giordano Resti A, Politi LS, Doglioni C, Cavalli F, Zucca E, Ponzoni M, Dolcetti R. Chlamydophila psittaci eradication with doxycycline as first-line targeted therapy for ocular adnexae lymphoma: final results of an international phase Ⅱ trial[J]. J Clin Oncol, 2012, 30(24): 2988-2994.
[DOI]
|
| [46] |
Fassi Fehri L, Mak TN, Laube B, Brinkmann V, Ogilvie LA, Mollenkopf H, Lein M, Schmidt T, Meyer TF, Brüggemann H. Prevalence of Propionibacterium acnes in diseased prostates and its inflammatory and transforming activity on prostate epithelial cells[J]. Int J Med Microbiol, 2011, 301(1): 69-78.
[DOI]
|
| [47] |
Shinohara DB, Vaghasia AM, Yu SH, Mak TN, Brüggemann H, Nelson WG, De Marzo AM, Yegnasubramanian S, Sfanos KS. A mouse model of chronic prostatic inflammation using a human prostate cancer-derived isolate of Propionibacterium acnes[J]. Prostate, 2013, 73(9): 1007-1015.
[DOI]
|
| [48] |
Quigley EMM. Gut microbiome as a clinical tool in gastrointestinal disease management: are we there yet?[J]. Nat Rev Gastroenterol Hepatol, 2017, 14(5): 315-320.
[DOI]
|
| [49] |
Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, Honda K, Ishikawa Y, Hara E, Ohtani N. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome[J]. Nature, 2013, 499(7456): 97-101.
[DOI]
|
| [50] |
Quante M, Bhagat G, Abrams JA, Marache F, Good P, Lee MD, Lee Y, Friedman R, Asfaha S, Dubeykovskaya Z, Mahmood U, Figueiredo JL, Kitajewski J, Shawber C, Lightdale CJ, Rustgi AK, Wang TC. Bile acid and inflammation activate gastric cardia stem cells in a mouse model of Barrett-like metaplasia[J]. Cancer Cell, 2012, 21(1): 36-51.
[DOI]
|
| [51] |
Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R, Lefkowitch JH, Bower M, Friedman R, Sartor RB, Rabadan R, Schwabe RF. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4[J]. Cancer Cell, 2012, 21(4): 504-516.
[DOI]
|
| [52] |
Goodman B, Gardner H. The microbiome and cancer[J]. J Pathol, 2018, 244(5): 667-676.
[DOI]
|
| [53] |
Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K, Thaiss CA, Reuben A, Livny J, Avraham R, Frederick DT, Ligorio M, Chatman K, Johnston SE, Mosher CM, Brandis A, Fuks G, Gurbatri C, Gopalakrishnan V, Kim M, Hurd MW, Katz M, Fleming J, Maitra A, Smith DA, Skalak M, Bu J, Michaud M, Trauger SA, Barshack I, Golan T, Sandbank J, Flaherty KT, Mandinova A, Garrett WS, Thayer SP, Ferrone CR, Huttenhower C, Bhatia SN, Gevers D, Wargo JA, Golub TR, Straussman R. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine[J]. Science, 2017, 357(6356): 1156-1160.
[DOI]
|
| [54] |
Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, Poirier-Colame V, Roux A, Becharef S, Formenti S, Golden E, Cording S, Eberl G, Schlitzer A, Ginhoux F, Mani S, Yamazaki T, Jacquelot N, Enot DP, Bérard M, Nigou J, Opolon P, Eggermont A, Woerther PL, Chachaty E, Chaput N, Robert C, Mateus C, Kroemer G, Raoult D, Boneca IG, Carbonnel F, Chamaillard M, Zitvogel L. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota[J]. Science, 2015, 350(6264): 1079-1084.
[DOI]
|
| [55] |
Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, Chang EB, Gajewski TF. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy[J]. Science, 2015, 350(6264): 1084-1089.
[DOI]
|
| [56] |
Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, Cogdill AP, Zhao L, Hudgens CW, Hutchinson DS, Manzo T, Petaccia de Macedo M, Cotechini T, Kumar T, Chen WS, Reddy SM, Szczepaniak Sloane R, Galloway-Pena J, Jiang H, Chen PL, Shpall EJ, Rezvani K, Alousi AM, Chemaly RF, Shelburne S, Vence LM, Okhuysen PC, Jensen VB, Swennes AG, McAllister F, Marcelo Riquelme Sanchez E, Zhang Y, Le Chatelier E, Zitvogel L, Pons N, Austin-Breneman JL, Haydu LE, Burton EM, Gardner JM, Sirmans E, Hu J, Lazar AJ, Tsujikawa T, Diab A, Tawbi H, Glitza IC, Hwu WJ, Patel SP, Woodman SE, Amaria RN, Davies MA, Gershenwald JE, Hwu P, Lee JE, Zhang J, Coussens LM, Cooper ZA, Futreal PA, Daniel CR, Ajami NJ, Petrosino JF, Tetzlaff MT, Sharma P, Allison JP, Jenq RR, Wargo JA. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients[J]. Science, 2018, 359(6371): 97-103.
[DOI]
|
| [57] |
Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, Gajewski TF. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients[J]. Science, 2018, 359(6371): 104-108.
[DOI]
|
 2018, Vol. 13
2018, Vol. 13


