随着人类疾病谱的改变[1-2],乙型肝炎病毒(hepatitis B virus,HBV)与肝癌[3](hepatitis B virus-associated carcinoma)、幽门螺杆菌与胃癌[4](helicobacter pylori-associated gastric cancer)、爱泼斯坦-巴尔病毒(Epstein-Barr virus, EBV)与淋巴瘤[5](Epstein-Barr virus-associated lymphoma)以及卡波西肉瘤相关病毒(Kaposi's sarcoma-associated herpesvirus, KSHV)与卡波西肉瘤[6](Kaposi's sarcoma)等慢性感染相关肿瘤的发生,逐渐成为全球人类健康的重要威胁因素。但是,国内外对于肿瘤的研究一直缺乏有效的体外研究模型[7],更是缺乏针对病原体感染所引发肿瘤的模型。类器官(organoid)作为一种新型的体外三维(three dimensions,3D)技术培养的组织器官模型,无疑为这一领域的研究拓宽了思路。类器官不仅能够高度模拟原位组织在体内的结构和功能,在基因水平上维持其稳定性,还可以弥补常规构建的人源组织肿瘤细胞系不能维持肿瘤异质性,以及人源组织肿瘤异种移植效率低、周期长等缺点[7-26]。随着研究技术的不断完善和成熟,越来越多数据表明了类器官组织已在多种疾病和应用研究领域(包括感染肿瘤致病机制、药物试验、个体化治疗等方面)发挥重要作用[7-26]。因此,本文针对类器官组织在感染性疾病模型和应用研究领域的进展进行综述,期望对读者了解该领域的研究动态有所帮助。
1 类器官发展概况类器官是一种在体外环境下培育而成的具有3D结构的“迷你器官”(mini-organs),拥有类似真实器官的复杂结构,并能最大限度模拟来源组织或器官的生理功能,同时还可以在体外进行传代和冻存。除了正常组织的干细胞能够在体外3D培养生长为类器官外,肿瘤组织也可经体外3D培养生长为肿瘤类器官。目前,类器官3D培养技术已经涉及多种人体多种组织器官,如胰腺、脑、视网膜、肝、脾、前列腺和肺等(见图 1)[7-26]。类器官的概念最早于1946年由Smith和Cochrane[27]在皮样囊肿(dermoid cysts)的研究中提出,但直到2014年Lancaster和Knoblich[28]才明确定义了类器官是以干细胞或器官组织细胞为原料,经体外培养分化后自组装为器官样结构的细胞群。在类器官发展进程中最重要的里程碑发现是,2009年Clevers团队将小肠上皮绒毛-隐窝(crypt-villus)基底部来源的Lgr5+干细胞(Lgr5+ stem cells)从小鼠体内分离处理后加到含有生长因子的基质胶中,Lgr5+干细胞能够在3D基质胶的支撑下,生长分化成为中空的囊腔状球体类器官,高度模拟小肠上皮在体内的结构和功能以及基因的稳定性[7, 29]。在此项研究的基础上,通过前期摸索,本课题组也已成功建立了肠道类器官的培养体系(见图 2)。同年Gao等[30]从临床前列腺肿瘤患者活检组织中亦成功地培育出人前列腺肿瘤类器官,并证实肿瘤类器官可用于体外药物检测与体内异种移植。

|
| 图 1 不同组织类器官的发展历程 Fig. 1 The development timeline of organoid from different tissues |
|
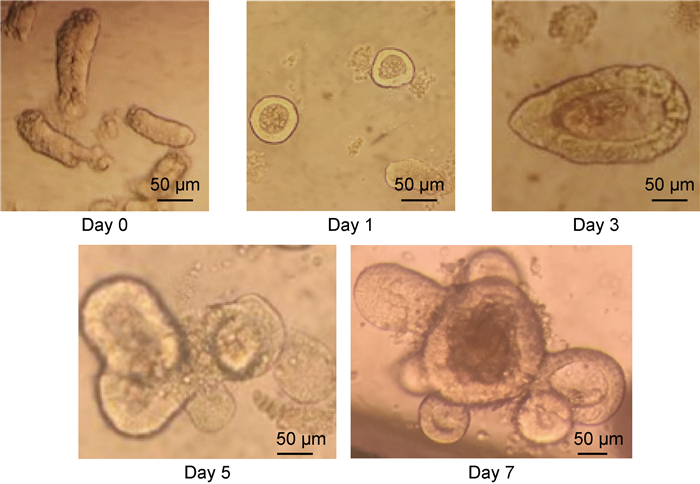
| The crypts were isolated from adult mouse intestine, and grown as three-dimensional spheroids in matrigel covered by 50% conditioned medium collected from the supplement of L-WRN cells. Crypts formed spheroid structures on day 1 and could bud on day 5. 图 2 体外3D技术培养的小鼠小肠类器官 Fig. 2 The progress of mouse intestinal organoids 3D culture in vitro |
2016年Nozaki等[31]建立了小肠类器官与小肠上皮T细胞共培养体系,发现类器官微环境可以提高T细胞体外的生存时间,而且T细胞的运动轨迹与在动物体内的相似。根据该研究,本课题组也成功建立了小肠类器官与T淋巴细胞共培养体系(见图 3)。随后,Dijkstra等[32]通过将人肿瘤类器官与自身外周血淋巴细胞共培养,发现共培养可以从结肠癌和非小细胞肺癌患者外周血中富集肿瘤反应性T细胞,并证明富集的反应性T细胞可用来评估肿瘤类器官的杀伤效果,进一步推动了肿瘤免疫治疗的发展。
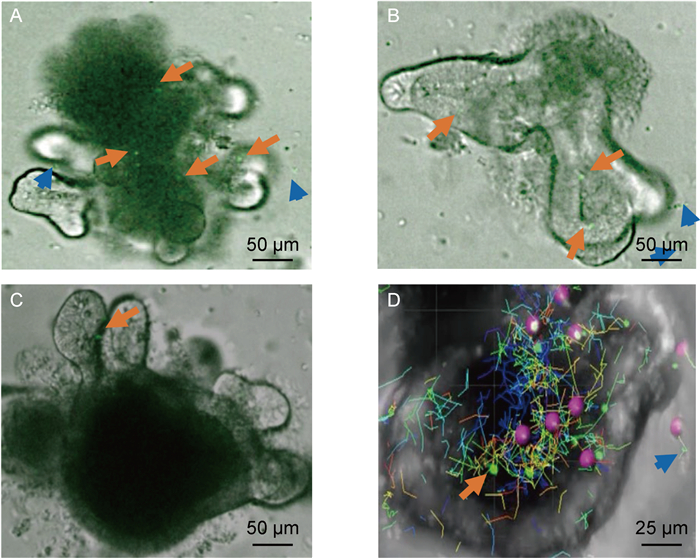
|
| Intestinal organoids were cultured for 2 days prior to co-culture with T cells. On the day of co-culture, T cells with GFP were isolated from EGFP transgenic mice, and organoids were released from matrigel. 100 organoids and 1.0×105 T cells were mixed with DMEM and placed together onto a 24-well plate to solidify at the incubator for 30 mins and covered by 50% conditioned medium with IL-2, IL-7 and IL-15. The medium was refreshed every 2 days and image after co-culture 48 h. The data was analysed by Imaris software, and the long orange arrows show the T cells entrance into the organoids, the short blue arrows show the T cells outside of the organoids. In picture D, the red balls stand for the T cells with GFP in Imaris software and the different lines show the moving path of T cells. 图 3 小肠类器官与T细胞共培养模型 Fig. 3 Co-culture intestinal organoids and T cells |
2019年Wimmer等[26]首次从多功能干细胞中成功培育出包含有内皮细胞与管周细胞的人体血管类器官。将人体血管类器官移植到小鼠体内,发现95%的人血管类器官能够在小鼠体内存活并生长达6个月之久,血管类器官在小鼠体内能正常发育成功,有完善的动脉、静脉及毛细血管。通过对人血管类器官移植小鼠进行药物处理诱导糖尿病后发现,人体血管类器官基底膜增厚,模拟了糖尿病患者的微血管变化。人血管类器官能够再现人类血管的结构与功能,为血管疾病和糖尿病等多种基础性疾病的防控提供了新的思路。同年,Cakir等[33]在研究脑类器官时,在培养的人皮质类器官(human cortical organoids,hCOs)中表达ETV2转录因子,成功形成具有微血管网络结构和类血-脑屏障结构的血管化人皮质类器官(vascularized hCOs,vhCOs)。该项研究不仅提高了脑类器官的培养成熟度和分化程度,还克服了类器官缺乏血管网络系统的弊端。血管类器官和具有血管结构脑类器官的研究成为类器官研究的又一个重要里程碑。
随着类器官研究技术的不断成熟,类器官血管形成、神经支配及与免疫细胞共培养等技术瓶颈的不断突破,类器官应用范围将越来越广泛。
2 感染性疾病及其研究模型 2.1 感染性疾病近年来,癌症严重危害着人类的身心健康,已成为世界范围内死亡率显著上升的一个重要原因。2017年全球十大癌症发病排名指出,结肠癌和直肠癌(colon and rectum cancer)位居第4,乳腺癌(breast cancer)位居第3,气管、支气管和肺癌(tracheal, bronchus, and lung cancer)位居第2,非黑色素皮肤癌(nonmelanoma skin cancer)位居第1,此外胃癌(stomach cancer)、肝癌(liver cancer)等发病率亦居高不下[34]。预计到2025年,每年将有1 930万新发癌症病例,并且一半以上的癌症病例和死亡病例将发生在中低收入国家[35-36]。2019年9月,Christina等报道,癌症和其他非传染性疾病已成为全球健康发展的主要威胁,并预计癌症将成为21世纪人类因疾病死亡的主要原因和增加预期寿命的最主要障碍[34, 37],该声明进一步强调了深入研究癌症对于世界健康发展的重要性。
引起癌症发生、发展的因素是多方面的,包括外在物理因素(电辐射、紫外线等),化学因素(亚硝胺类、黄曲霉素等),不良的生活习惯(吸烟、饮酒、熬夜等),遗传因素(基因突变或缺失等)以及病原体感染等。研究表明,99%的人在其一生中至少被一种疾病相关病原感染,但是癌症发生、发展的很多具体机制至今尚不清楚[34, 37]。由病原体感染所致的肿瘤又被称为感染性肿瘤,据研究报道,至少约有15%~20%的癌症与病毒感染相关,如人乳头瘤病毒(human papilloma virus,HPV)与宫颈癌,HBV/丙型肝炎病毒(hepatitis C virus, HCV)与肝癌[38]。第1个被证实的感染性肿瘤是1911年由Rous[39]发现的由致瘤病毒——劳斯肉瘤病毒(Rous sarcoma virus,RSV)感染引发禽类的劳斯肉瘤(Rous sarcoma)。Epstein等[40]在Burkitt淋巴瘤细胞培养液中发现人类第1个致瘤性病毒——EB病毒,该病毒被证实与多种淋巴瘤及鼻咽癌密切相关。随后,HBV与肝癌,HPV与宫颈癌,人类嗜T淋巴细胞病毒-1(human T-cell lymphotropic virus type-1, HTLV-1)与成人T细胞白血病,人类免疫缺陷病毒(human immunodeficiency virus,HIV)与艾滋病,KSHV与卡波西肉瘤等病毒与癌症的相互作用关系被先后报道,进一步证实感染在疾病发生中的重要作用[41]。
引发感染性疾病的病原不仅包含病毒,还有多种细菌与寄生虫。幽门螺杆菌(Helicobacter pylori,Hp)是第1个被证实能够引发胃炎及胃癌的细菌[42-46]。除此之外,研究还发现梭形杆菌、卟琳单胞菌、链球菌等与结肠癌相关,伤寒沙门菌与胆囊癌相关[47],巴尔通体与血管瘤相关[48]。寄生虫引发疾病的研究也逐渐成熟,现已证实肝吸虫能引起胆管癌,血吸虫能引发膀胱癌,阴道毛滴虫与弓形体分别与前列腺癌和脑肿瘤形成相关[49-50]。
据世界卫生组织(World Health Organization, WHO)统计,感染人类的病原体中至少有12%会引起新发感染性疾病[51],包括由疟原虫感染引起的疟疾,严重急性呼吸综合征冠状病毒(severe acute respiratory syndrome coronavirus,SARS-CoV)引发的SARS, 禽流感病毒感染人引发的流感,中东呼吸综合征冠状病毒(Middle East respiratory syndrome coronavirus, MERS-CoV)引发的MERS,埃博拉病毒(Ebola virus)引发的出血热,寨卡病毒(Zika virus)感染所致的畸形及现在正在流行的由严重急性呼吸综合征冠状病毒2(severe acute respiratory syndrome coronavirus-2,SARS-CoV-2)引发的2019冠状病毒病(corona virus disease 2019,COVID-19)。这些病原感染所致的新发感染性疾病,由于发病急、传染性强,一旦发病就会迅速流行,因此,针对感染性疾病的研究尤为关键。
2.2 疾病研究模型目前在肿瘤研究领域较为常用的模型为2D水平的细胞模型、3D水平的类器官模型和动物模型。2D水平的细胞模型虽能在体外维持肿瘤的无限增殖与侵袭生长能力,但不能维持肿瘤体内的异质性(如临床上某些常见的癌基因突变,在细胞模型中并不存在)[52-53];动物模型虽能较好地维持肿瘤的异质性,但存在物种跨越的局限性,并对药物的反应性不稳定,且耗时耗力,还存在一定的伦理问题[54];而3D水平的类器官模型不仅可以维持肿瘤的异质性,高度模拟体内原位组织的生理结构和功能,还可以在基因水平进行基因编辑,操作简单方便,更利于观察组织细胞的生长、运动等情况。类器官模型很好地弥补了细胞模型与动物模型的缺陷,为感染性疾病的体外研究提供了更好的前景,可广泛应用于疾病模型、药物筛选等多方面的研究,其应用前景十分广阔,被誉为“朝阳研究”[55-56]。
3 类器官在病原感染性疾病研究中的应用因类器官与来源器官组织的基因、结构和功能的高度相似性,其已广泛应用于模拟体内组织细胞生长、分化和器官形成过程,以及药物筛选与评价、个性化治疗、生物医学材料及组织工程等方面。随着病原体感染与肿瘤发生关系研究的深入,感染性疾病已成为肿瘤研究领域的焦点问题。作为新型的研究模型,3D技术培养的类器官结合显微注射技术能更好地模拟宿主与病原体之间的相互作用,将为研究疾病的发生、发展、结局、转归,药物选择,个体化治疗以及精准医学等领域提供新的方向[57]。现已有多种类器官作为疾病感染模型。
3.1 消化道类器官胃肠道类器官研究较为成熟,其感染模型应用也较为广泛。Bartfeld和Clevers[58]从人胃组织中分离出胃腺体,经处理后加入基质胶中并添加必要生长因子如表皮生长因子(epidermal growth factor,EGF)、R-spondin和Noggin等,成功在体外培养出胃类器官,并且利用显微注射技术,将幽门螺杆菌注入胃类器官中,观察幽门螺杆菌在胃类器官中定植的过程;Jiao等[59]用沙门菌感染小鼠小肠类器官,发现沙门菌的效应蛋白AvrA通过抑制自噬关键蛋白Beclin-1的表达,从而抑制自噬并促进细菌入侵宿主细胞进而在宿主内定植。Saxena等[60]收集不同患者肠道组织经体外培养人肠道类器官后,通过人轮状病毒感染实验,发现病毒能够引起类器官生理性管腔扩张。以上实验模拟了轮状病毒引起生理性腹泻的特征,从细胞水平研究肠道病毒—宿主的相互作用,为人类轮状病毒和其他肠道病原体(如人类诺如病毒、星形病毒和圆环病毒以及寄生虫和细菌)致病机制的研究提供了可行的感染模型。Drummond等[61]发现埃可病毒11型(Echovirus 11,E11)、柯萨奇病毒B组(Coxsachie virus B,CVB)和肠道病毒71型(enterovirus 71,EV71)等肠道病毒能够感染肠道类器官,激活炎症信号通路诱发抗病毒反应,进一步佐证肠道类器官能够高度模拟原位肠道的免疫防御和抗病原体保护功能。Ettayebi等[62]通过人诺如病毒HuNov感染肠道类器官建立肠道感染模型,并通过这种肠道类器官模型研究HuNov感染复制与组织血型抗原表达之间的作用关系,为其他肠道病毒与宿主相互作用研究提供了参考。2017年,Zhou等[63]利用MERS-CoV感染小肠类器官,发现小肠类器官对MERS-CoV高度易感,为MERS-CoV致病机制的研究提供有效的研究模型。Lamer等[64]用4种不同的方法培养小肠类器官,并用SARS-CoV与SARS-CoV-2分别进行感染,通过激光共聚焦显微镜观察类器官中被感染细胞的数量,发现在感染24 h可见单个感染的细胞,感染60 h内被感染细胞数量逐渐增加,随后对感染60 h的类器官进行电子显微镜观察,发现在类器官管腔及基底细胞内出现80~120 nm的病毒颗粒,表明小肠类器官可作为研究冠状病毒感染的新实验模型。
3.2 呼吸道类器官Zhou等[65]利用肺组织中的成体干细胞培养出3D生长的气道类器官,经诱导分化后成功模拟了自然情况下流感病毒感染人类呼吸道的过程,为人类呼吸系统疾病提供了一个崭新的研究平台。2019年Sachs等[25]从呼吸道患者支气管肺泡切除或灌洗材料中培养出气道类器官,通过呼吸道合胞病毒(respiratory syncytial virus, RSV)感染气道类器官实验发现,病毒能够利用其NS2非结构蛋白增强类器官的活力;同年,Porotto等[66]利用人多能干细胞培养肺支气管类器官,建立儿童及婴幼儿末梢肺组织副流感病毒感染模型,为呼吸道病毒感染及宿主病原相互作用关系研究提供了更好的模型。
3.3 脑类器官Garcez等[67]用诱导性多功能干细胞(induced pluripotent stem cell,iPSC)培养人脑类器官,并用寨卡病毒和登革病毒2型(dengue virus type 2,DENV-2)分别感染人大脑类器官,发现寨卡病毒感染脑类器官后可以抑制类器官的生长,抑制效率高达40%,与临床上寨卡病毒感染引起的儿童先天性小脑畸形的结果一致,而DENV-2感染并未引起类器官生长抑制。Zhang等[68]从人胚胎干细胞培养出端脑皮质类器官,该类器官具有多层前体区和所有6种神经元亚型,与人体内皮质发育相似。通过日本脑炎病毒(Japanese encephalitis virus, JEV)感染端脑皮质类器官实验研究JEV与日本脑炎发生、发展的关系,发现JEV感染能够促进脑类器官细胞死亡,导致类器官体积变小。
3.4 肝胆类器官Huch等[69]通过培养小鼠肝脏类器官进行HBV和HCV感染实验,研究病毒与肝炎、肝硬化及肝癌间的相互作用;在研究沙门菌与胆囊癌发生发展关系时,Scanu等[19]建立沙门菌感染鼠胆囊类器官模型,发现沙门菌感染可以诱导胆囊类器官TP53变异、C-myc扩增,从分子水平证实沙门菌是引起胆囊癌变的重要因素。在SARS-CoV-2引发COVID-19的关键时刻,Zhao等[70]应用人肝脏胆管类器官建立SARS-CoV-2感染模型,发现感染24 h后可检测到SARS-CoV-2的核衣壳蛋白(nucleocapsid protein,N protein),在胆管类器官内SARS-CoV-2载量显著增加,但是感染48 h后病毒载量下降。同时,上述研究发现SARS-CoV-2感染降低了类器官中Claudin 1的表达,并下调两个关键胆汁酸转运蛋白的表达(顶端钠离子/胆汁酸转运体与囊性纤维化穿膜传导调节蛋白),揭示SARS-CoV-2感染引发患者胆管功能紊乱和胆汁淤积,进而导致肝脏损伤。
3.5 血管类器官Monteil等[71]通过iPSC成功培养出人毛细血管类器官,并用临床分离的SARS-CoV-2毒株感染该血管类器官,感染3 d和6 d后用定量反转录聚合酶链反应(quantitative reverse transcriptase polymerase chain reaction,qRT-PCR)检测病毒RNA,发现感染6 d的病毒RNA载量明显高于感染3 d的病毒载量。SARS-CoV-2感染血管类器官6 d后的培养上清液能够再次感染Vero E6细胞,表明病毒在类器官中可以增殖并产生具有感染能力的子代病毒。
3.6 肾脏类器官血管紧张素转化酶2(angiotensin converting enzyme 2,ACE2)能够促进血管紧张素的成熟,在肺、心脏、肾脏和肠道中广泛存在,是SARS-CoV-2感染进入人体细胞的重要受体。Monteil等[71]从人胚胎干细胞培养出肾脏类器官,并发现肾类器官中含有表达ACE2的细胞簇。通过临床分离的SARS-CoV-2毒株感染肾类器官建立了病毒感染模型,加入人源可溶性重组ACE2(hrsACE2)能够有效抑制SARS-CoV-2在肾脏类器官中的增殖,抑制效果呈现剂量依赖性,结果表明hrsACE2可以抑制SARS-CoV-2增殖并阻断病毒进入宿主细胞,为COVID-19的治疗与防控提供了新的方向。
4 结语与展望类器官是利用体外3D技术培养干细胞而形成的多功能细胞团,具有自身增殖和多向分化的能力[72],能够高度模拟原位组织的结构和功能,已广泛应用于疾病建模、基因编辑、高通量药物筛选、毒性试验、新型治疗方法的开发、再生医学等方面,成为研究人类发育过程及疾病机制的有效平台[25, 73]。尽管如此,类器官在感染性疾病研究中的应用尚处于初级阶段,仍存在应用的局限性及一些亟待解决的问题:由于培养类器官所用的基质胶来源于动物肉瘤组织,可能会对细胞性质及药物筛选过程中的反应性产生未知的影响,限制了类器官在人体器官移植中的应用;iPSC衍生的类器官多数需要饲养层细胞,这增加了培养过程的复杂程度;类器官成像也是科学研究常面临的问题,高通量深度扫描成像能够改善类器官成像效果;大部分类器官缺乏血液、血管、淋巴管、免疫细胞和外周血管神经等基质成分,器官功能不成熟,不能很好地模拟疾病的发生过程[74];类器官的体积大小是限制其发展的关键因素,而类器官内氧与营养成分扩散的程度取决于其大小,缺氧及营养物质缺乏导致的类器官死亡是亟须解决的问题[75]。尽管类器官研究中存在这些限制,但这一技术的搭建实现了从基础到临床的快速转化,为感染性疾病尤其是新发传染性疾病致病机制研究、靶向药物筛选及个性化治疗提供了新的平台,将广泛应用于基础与临床领域的相关研究。
| [1] |
Marchevsky AM, Bottone EJ, Geller SA, Giger DK. The changing spectrum of disease, etiology, and diagnosis of mucormycosis[J]. Hum Pathol, 1980, 11(5): 457-464.
[DOI]
|
| [2] |
Hou JH, Zhu HX, Zhou ML, Le WB, Zeng CH, Liang SS, Xu F, Liang DD, Shao SJ, Liu Y, Liu ZH. Changes in the spectrum of kidney diseases: an analysis of 40, 759 biopsy-proven cases from 2003 to 2014 in China[J]. Kidney Dis (Basel), 2018, 4(1): 10-19.
[DOI]
|
| [3] |
Wu T, Zheng X, Yang M, Zhao A, Li M, Chen T, Panee J, Jia W, Ji G. Serum lipid alterations identified in chronic hepatitis B, hepatitis B virus-associated cirrhosis and carcinoma patients[J]. Sci Rep, 2017, 7: 42710.
[DOI]
|
| [4] |
Toyoda T, Tsukamoto T, Yamamoto M, Ban H, Saito N, Takasu S, Shi L, Saito A, Ito S, Yamamura Y, Nishikawa A, Ogawa K, Tanaka T, Tatematsu M. Gene expression analysis of a Helicobacter pylori-infected and high-salt diet-treated mouse gastric tumor model: identification of CD177 as a novel prognostic factor in patients with gastric cancer[J]. BMC Gastroenterol, 2013, 13: 122.
[DOI]
|
| [5] |
Vockerodt M, Yap LF, Shannon-Lowe C, Curley H, Wei W, Vrzalikova K, Murray PG. The Epstein-Barr virus and the pathogenesis of lymphoma[J]. J Pathol, 2015, 235(2): 312-322.
[DOI]
|
| [6] |
Goncalves PH, Uldrick TS, Yarchoan R. HIV-associated Kaposi sarcoma and related diseases[J]. AIDS, 2017, 31(14): 1903-1916.
[DOI]
|
| [7] |
Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, Van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche[J]. Nature, 2009, 459(7244): 262-265.
[DOI]
|
| [8] |
Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium[J]. Gastroenterology, 2011, 141(5): 1762-1772.
[DOI]
|
| [9] |
Peloso A, Katari R, Patel T, Hemal S, Zambon JP, Salvatori M, Orlando G. Considerations on the development of a model of kidney bioengineering and regeneration in rats[J]. Expert Rev Med Devices, 2013, 10(5): 597-601.
[DOI]
|
| [10] |
Willenbring H, Soto-Gutierrez A. Transplantable liver organoids made from only three ingredients[J]. Cell Stem Cell, 2013, 13(2): 139-140.
[DOI]
|
| [11] |
Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly[J]. Nature, 2013, 501(7467): 373-379.
[DOI]
|
| [12] |
Chua CW, Shibata M, Lei M, Toivanen R, Barlow LJ, Bergren SK, Badani KK, Mckiernan JM, Benson MC, Hibshoosh H, Shen MM. Single luminal epithelial progenitors can generate prostate organoids in culture[J]. Nat Cell Biol, 2014, 16(10): 951-961.
[DOI]
|
| [13] |
Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel H, Sachs N, Vries RGJ, Cuppen E, Chen Y, Sawyers CL, Clevers HC. Identification of multipotent luminal progenitor cells in human prostate organoid cultures[J]. Cell, 2014, 159(1): 163-175.
[DOI]
|
| [14] |
Deward AD, Cramer J, Lagasse E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population[J]. Cell Rep, 2014, 9(2): 701-711.
[DOI]
|
| [15] |
Ren W, Lewandowski BC, Watson J, Aihara E, Iwatsuki K, Bachmanov AA, Margolskee RF, Jiang P. Single Lgr5-or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo[J]. Proc Natl Acad Sci U S A, 2014, 111(46): 16401-16406.
[DOI]
|
| [16] |
Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, Ellis E, van Wenum M, Fuchs SA, de Ligt J, van de Wetering M, Sasaki N, Boers SJ, Kemperman H, de Jonge J, Ijzermans JN, Nieuwenhuis EE, Hoekstra R, Strom S, Vries RR, van der Laan LJ, Cuppen E, Clevers H. Long-term culture of genome-stable bipotent stem cells from adult human liver[J]. Cell, 2015, 160(1/2): 299-312.
[DOI]
|
| [17] |
Kessler M, Hoffmann K, Brinkmann V, Thieck O, Jackisch S, Toelle B, Berger H, Mollenkopf HJ, Mangler M, Sehouli J, Fotopoulou C, Meyer TF. The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids[J]. Nat Commun, 2015, 6: 8989.
[DOI]
|
| [18] |
Boj SF, Hwang CI, Baker LA, Chio Ⅱ, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, Gracanin A, Oni T, Yu KH, Van Boxtel R, Huch M, Rivera KD, Wilson JP, Feigin ME, Ohlund D, Handly-Santana A, Ardito-Abraham CM, Ludwig M, Elyada E, Alagesan B, Biffi G, Yordanov GN, Delcuze B, Creighton B, Wright K, Park Y, Morsink FH, Molenaar IQ, Borel Rinkes IH, Cuppen E, Hao Y, Jin Y, Nijman IJ, Iacobuzio-Donahue C, Leach SD, Pappin DJ, Hammell M, Klimstra DS, Basturk O, Hruban RH, Offerhaus GJ, Vries RG, Clevers H, Tuveson DA. Organoid models of human and mouse ductal pancreatic cancer[J]. Cell, 2015, 160(1/2): 324-338.
|
| [19] |
Scanu T, Spaapen RM, Bakker JM, Pratap CB, Wu LE, Hofland I, Broeks A, Shukla VK, Kumar M, Janssen H, Song JY, Neefjes-Borst EA, te Riele H, Holden DW, Nath G, Neefjes J. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma[J]. Cell Host Microbe, 2015, 17(6): 763-774.
[DOI]
|
| [20] |
Maimets M, Rocchi C, Bron R, Pringle S, Kuipers J, Giepmans BN, Vries RG, Clevers H, de Haan G, van Os R, Coppes RP. Long-term in vitro expansion of salivary gland stem cells driven by Wnt signals[J]. Stem Cell Reports, 2016, 6(1): 150-162.
[DOI]
|
| [21] |
Longworth-Mills E, Koehler KR, Hashino E. Generating inner ear organoids from mouse embryonic stem cells[J]. Methods Mol Biol, 2016, 1341: 391-406.
[DOI]
|
| [22] |
Völkner M, Zschätzsch M, Rostovskaya M, Overall RW, Busskamp V, Anastassiadis K, Karl MO. Retinal organoids from pluripotent stem cells efficiently recapitulate retinogenesis[J]. Stem Cell Reports, 2016, 6(4): 525-538.
[DOI]
|
| [23] |
Carter EP, Gopsill JA, Gomm JJ, Jones JL, Grose RP. A 3D in vitro model of the human breast duct: a method to unravel myoepithelial-luminal interactions in the progression of breast cancer[J]. Breast Cancer Res, 2017, 19(1): 50.
[DOI]
|
| [24] |
Kijima T, Nakagawa H, Shimonosono M, Chandramouleeswaran PM, Hara T, Sahu V, Kasagi Y, Kikuchi O, Tanaka K, Giroux V, Muir AB, Whelan KA, Ohashi S, Naganuma S, Klein-Szanto AJ, Shinden Y, Sasaki K, Omoto I, Kita Y, Muto M, Bass AJ, Diehl JA, Ginsberg GG, Doki Y, Mori M, Uchikado Y, Arigami T, Avadhani NG, Basu D, Rustgi AK, Natsugoe S. Three-dimensional organoids reveal therapy resistance of esophageal and oropharyngeal squamous cell carcinoma cells[J]. Cell Mol Gastroenterol Hepatol, 2019, 7(1): 73-91.
[DOI]
|
| [25] |
Sachs N, Papaspyropoulos A, Zomer-Van Ommen DD, Heo I, Bottinger L, Klay D, Weeber F, Huelsz-Prince G, Iakobachvili N, Amatngalim GD, De Ligt J, Van Hoeck A, Proost N, Viveen MC, Lyubimova A, Teeven L, Derakhshan S, Korving J, Begthel H, Dekkers JF, Kumawat K, Ramos E, Van Oosterhout MF, Offerhaus GJ, Wiener DJ, Olimpio EP, Dijkstra KK, Smit EF, Van Der Linden M, Jaksani S, Van De Ven M, Jonkers J, Rios AC, Voest EE, Van Moorsel CH, Van Der Ent CK, Cuppen E, Van Oudenaarden A, Coenjaerts FE, Meyaard L, Bont LJ, Peters PJ, Tans SJ, Van Zon JS, Boj SF, Vries RG, Beekman JM, Clevers H. Long-term expanding human airway organoids for disease modeling[J]. EMBO J, 2019, 38(4): e100300.
[DOI]
|
| [26] |
Wimmer RA, Leopoldi A, Aichinger M, Wick N, Hantusch B, Novatchkova M, Taubenschmid J, Hämmerle M, Esk C, Bagley JA, Lindenhofer D, Chen G, Boehm M, Agu CA, Yang F, Fu B, Zuber J, Knoblich JA, Kerjaschki D, Penninger JM. Human blood vessel organoids as a model of diabetic vasculopathy[J]. Nature, 2019, 565(7740): 505-510.
[DOI]
|
| [27] |
Smith E, Cochrane WJ. Cystic organoid teratoma: (report of a case)[J]. Can Med Assoc J, 1946, 55(2): 151-152.
[PubMed]
|
| [28] |
Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies[J]. Science, 2014, 345(6194): 1247125.
[DOI]
|
| [29] |
Li X, Francies HE, Secrier M, Perner J, Miremadi A, Galeano-Dalmau N, Barendt WJ, Letchford L, Leyden GM, Goffin EK, Barthorpe A, Lightfoot H, Chen E, Gilbert J, Noorani A, Devonshire G, Bower L, Grantham A, Macrae S, Grehan N, Wedge DC, Fitzgerald RC, Garnett MJ. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics[J]. Nat Commun, 2018, 9(1): 2983.
[DOI]
|
| [30] |
Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK, Wongvipat J, Kossai M, Ramazanoglu S, Barboza LP, Di W, Cao Z, Zhang QF, Sirota I, Ran L, Macdonald TY, Beltran H, Mosquera JM, Touijer KA, Scardino PT, Laudone VP, Curtis KR, Rathkopf DE, Morris MJ, Danila DC, Slovin SF, Solomon SB, Eastham JA, Chi P, Carver B, Rubin MA, Scher HI, Clevers H, Sawyers CL, Chen Y. Organoid cultures derived from patients with advanced prostate cancer[J]. Cell, 2014, 159(1): 176-187.
[DOI]
|
| [31] |
Nozaki K, Mochizuki W, Matsumoto Y, Matsumoto T, Fukuda M, Mizutani T, Watanabe M, Nakamura T. Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes[J]. J Gastroenterol, 2016, 51(3): 206-213.
[DOI]
|
| [32] |
Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, Slagter M, van der Velden DL, Kaing S, Kelderman S, van Rooij N, van Leerdam ME, Depla A, Smit EF, Hartemink KJ, de Groot R, Wolkers MC, Sachs N, Snaebjornsson P, Monkhorst K, Haanen J, Clevers H, Schumacher TN, Voest EE. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids[J]. Cell, 2018, 174(6): 1586-1598.
[DOI]
|
| [33] |
Cakir B, Xiang Y, Tanaka Y, Kural MH, Parent M, Kang YJ, Chapeton K, Patterson B, Yuan Y, He CS, Raredon MSB, Dengelegi J, Kim KY, Sun P, Zhong M, Lee S, Patra P, Hyder F, Niklason LE, Lee SH, Yoon YS, Park IH. Engineering of human brain organoids with a functional vascular-like system[J]. Nat Methods, 2019, 16(11): 1169-1175.
[DOI]
|
| [34] |
Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Abate D, Abbasi N, Abbastabar H, Abd-Allah F, Abdel-Rahman O, Abdelalim A, Abdoli A, Abdollahpour I, Abdulle ASM, Abebe ND, Abraha HN, Abu-Raddad LJ, Abualhasan A, Adedeji IA, Advani SM, Afarideh M, Afshari M, Aghaali M, Agius D, Agrawal S, Ahmadi A, Ahmadian E, Ahmadpour E, Ahmed MB, Akbari ME, Akinyemiju T, Al-Aly Z, Alabdulkader AM, Alahdab F, Alam T, Alamene GM, Alemnew BTT, Alene KA, Alinia C, Alipour V, Aljunid SM, Bakeshei FA, Almadi MAH, Almasi-Hashiani A, Alsharif U, Alsowaidi S, Alvis-Guzman N, Amini E, Amini S, Amoako YA, Anbari Z, Anber NH, Andrei CL, Anjomshoa M, Ansari F, Ansariadi A, Appiah SCY, Arab-Zozani M, Arabloo J, Arefi Z, Aremu O, Areri HA, Artaman A, Asayesh H, Asfaw ET, Ashagre AF, Assadi R, Ataeinia B, Atalay HT, Ataro Z, Atique S, Ausloos M, Avila-Burgos L, Avokpaho E, Awasthi A, Awoke N, Ayala Quintanilla BP, Ayanore MA, Ayele HT, Babaee E, Bacha U, Badawi A, Bagherzadeh M, Bagli E, Balakrishnan S, Balouchi A, Bärnighausen TW, Battista RJ, Behzadifar M, Behzadifar M, Bekele BB, Belay YB, Belayneh YM, Berfield KKS, Berhane A, Bernabe E, Beuran M, Bhakta N, Bhattacharyya K, Biadgo B, Bijani A, Bin Sayeed MS, Birungi C, Bisignano C, Bitew H, Bjørge T, Bleyer A, Bogale KA, Bojia HA, Borzì AM, Bosetti C, Bou-Orm IR, Brenner H, Brewer JD, Briko AN, Briko NI, Bustamante-Teixeira MT, Butt ZA, Carreras G, Carrero JJ, Carvalho F, Castro C, Castro F, Catalá-López F, Cerin E, Chaiah Y, Chanie WF, Chattu VK, Chaturvedi P, Chauhan NS, Chehrazi M, Chiang PP, Chichiabellu TY, Chido-Amajuoyi OG, Chimed-Ochir O, Choi JJ, Christopher DJ, Chu DT, Constantin MM, Costa VM, Crocetti E, Crowe CS, Curado MP, Dahlawi SMA, Damiani G, Darwish AH, Daryani A, Das Neves J, Demeke FM, Demis AB, Demissie BW, Demoz GT, Denova-Gutiérrez E, Derakhshani A, Deribe KS, Desai R, Desalegn BB, Desta M, Dey S, Dharmaratne SD, Dhimal M, Diaz D, Dinberu MTT, Djalalinia S, Doku DT, Drake TM, Dubey M, Dubljanin E, Duken EE, Ebrahimi H, Effiong A, Eftekhari A, El Sayed I, Zaki MES, El-Jaafary SI, El-Khatib Z, Elemineh DA, Elkout H, Ellenbogen RG, Elsharkawy A, Emamian MH, Endalew DA, Endries AY, Eshrati B, Fadhil I, Fallah V, Faramarzi M, Farhangi MA, Farioli A, Farzadfar F, Fentahun N, Fernandes E, Feyissa GT, Filip I, Fischer F, Fisher JL, Force LM, Foroutan M, Freitas M, Fukumoto T, Futran ND, Gallus S, Gankpe FG, Gayesa RT, Gebrehiwot TT, Gebremeskel GG, Gedefaw GA, Gelaw BK, Geta B, Getachew S, Gezae KE, Ghafourifard M, Ghajar A, Ghashghaee A, Gholamian A, Gill PS, Ginindza TTG, Girmay A, Gizaw M, Gomez RS, Gopalani SV, Gorini G, Goulart BNG, Grada A, Ribeiro Guerra M, Guimaraes ALS, Gupta PC, Gupta R, Hadkhale K, Haj-Mirzaian A, Haj-Mirzaian A, Hamadeh RR, Hamidi S, Hanfore LK, Haro JM, Hasankhani M, Hasanzadeh A, Hassen HY, Hay RJ, Hay SI, Henok A, Henry NJ, Herteliu C, Hidru HD, Hoang CL, Hole MK, Hoogar P, Horita N, Hosgood HD, Hosseini M, Hosseinzadeh M, Hostiuc M, Hostiuc S, Househ M, Hussen MM, Ileanu B, Ilic MD, Innos K, Irvani SSN, Iseh KR, Islam SMS, Islami F, Jafari Balalami N, Jafarinia M, Jahangiry L, Jahani MA, Jahanmehr N, Jakovljevic M, James SL, Javanbakht M, Jayaraman S, Jee SH, Jenabi E, Jha RP, Jonas JB, Jonnagaddala J, Joo T, Jungari SB, Jürisson M, Kabir A, Kamangar F, Karch A, Karimi N, Karimian A, Kasaeian A, Kasahun GG, Kassa B, Kassa TD, Kassaw MW, Kaul A, Keiyoro PN, Kelbore AG, Kerbo AA, Khader YS, Khalilarjmandi M, Khan EA, Khan G, Khang YH, Khatab K, Khater A, Khayamzadeh M, Khazaee-Pool M, Khazaei S, Khoja AT, Khosravi MH, Khubchandani J, Kianipour N, Kim D, Kim YJ, Kisa A, Kisa S, Kissimova-Skarbek K, Komaki H, Koyanagi A, Krohn KJ, Bicer BK, Kugbey N, Kumar V, Kuupiel D, La Vecchia C, Lad DP, Lake EA, Lakew AM, Lal DK, Lami FH, Lan Q, Lasrado S, Lauriola P, Lazarus JV, Leigh J, Leshargie CT, Liao Y, Limenih MA, Listl S, Lopez AD, Lopukhov PD, Lunevicius R, Madadin M, Magdeldin S, El Razek HMA, Majeed A, Maleki A, Malekzadeh R, Manafi A, Manafi N, Manamo WA, Mansourian M, Mansournia MA, Mantovani LG, Maroufizadeh S, Martini SMS, Mashamba-Thompson TP, Massenburg BB, Maswabi MT, Mathur MR, Mcalinden C, Mckee M, Meheretu HAA, Mehrotra R, Mehta V, Meier T, Melaku YA, Meles GG, Meles HG, Melese A, Melku M, Memiah PTN, Mendoza W, Menezes RG, Merat S, Meretoja TJ, Mestrovic T, Miazgowski B, Miazgowski T, Mihretie KMM, Miller TR, Mills EJ, Mir SM, Mirzaei H, Mirzaei HR, Mishra R, Moazen B, Mohammad DK, Mohammad KA, Mohammad Y, Darwesh AM, Mohammadbeigi A, Mohammadi H, Mohammadi M, Mohammadian M, Mohammadian-Hafshejani A, Mohammadoo-Khorasani M, Mohammadpourhodki R, Mohammed AS, Mohammed JA, Mohammed S, Mohebi F, Mokdad AH, Monasta L, Moodley Y, Moosazadeh M, Moossavi M, Moradi G, Moradi-Joo M, Moradi-Lakeh M, Moradpour F, Morawska L, Morgado-Da-Costa J, Morisaki N, Morrison SD, Mosapour A, Mousavi SM, Muche AA, Muhammed OSS, Musa J, Nabhan AR, Naderi M, Nagarajan AJ, Nagel G, Nahvijou A, Naik G, Najafi F, Naldi L, Nam HS, Nasiri N, Nazari J, Negoi I, Neupane S, Newcomb PA, Nggada HA, Ngunjiri JW, Nguyen CT, Nikniaz L, Ningrum DNA, Nirayo YL, Nixon MR, Nnaji CA, Nojomi M, Nosratnejad S, Shiadeh MN, Obsa MS, Ofori-Asenso R, Ogbo FA, Oh IH, Olagunju AT, Olagunju TO, Oluwasanu MM, Omonisi AE, Onwujekwe OE, Oommen AM, Oren E, Ortega-Altamirano DDV, Ota E, Otstavnov SS, Owolabi MO, P AM, Padubidri JR, Pakhale S, Pakpour AH, Pana A, Park EK, Parsian H, Pashaei T, Patel S, Patil ST, Pennini A, Pereira DM, Piccinelli C, Pillay JD, Pirestani M, Pishgar F, Postma MJ, Pourjafar H, Pourmalek F, Pourshams A, Prakash S, Prasad N, Qorbani M, Rabiee M, Rabiee N, Radfar A, Rafiei A, Rahim F, Rahimi M, Rahman MA, Rajati F, Rana SM, Raoofi S, Rath GK, Rawaf DL, Rawaf S, Reiner RC, Renzaho AMN, Rezaei N, Rezapour A, Ribeiro AI, Ribeiro D, Ronfani L, Roro EM, Roshandel G, Rostami A, Saad RS, Sabbagh P, Sabour S, Saddik B, Safiri S, Sahebkar A, Salahshoor MR, Salehi F, Salem H, Salem MR, Salimzadeh H, Salomon JA, Samy AM, Sanabria J, Santric Milicevic MM, Sartorius B, Sarveazad A, Sathian B, Satpathy M, Savic M, Sawhney M, Sayyah M, Schneider IJC, Schöttker B, Sekerija M, Sepanlou SG, Sepehrimanesh M, Seyedmousavi S, Shaahmadi F, Shabaninejad H, Shahbaz M, Shaikh MA, Shamshirian A, Shamsizadeh M, Sharafi H, Sharafi Z, Sharif M, Sharifi A, Sharifi H, Sharma R, Sheikh A, Shirkoohi R, Shukla SR, Si S, Siabani S, Silva DAS, Silveira DGA, Singh A, Singh JA, Sisay S, Sitas F, Sobngwi E, Soofi M, Soriano JB, Stathopoulou V, Sufiyan MB, Tabarés-Seisdedos R, Tabuchi T, Takahashi K, Tamtaji OR, Tarawneh MR, Tassew SG, Taymoori P, Tehrani-Banihashemi A, Temsah MH, Temsah O, Tesfay BE, Tesfay FH, Teshale MY, Tessema GA, Thapa S, Tlaye KG, Topor-Madry R, Tovani-Palone MR, Traini E, Tran BX, Tran KB, Tsadik AG, Ullah I, Uthman OA, Vacante M, Vaezi M, Varona Pérez P, Veisani Y, Vidale S, Violante FS, Vlassov V, Vollset SE, Vos T, Vosoughi K, Vu GT, Vujcic IS, Wabinga H, Wachamo TM, Wagnew FS, Waheed Y, Weldegebreal F, Weldesamuel GT, Wijeratne T, Wondafrash DZ, Wonde TE, Wondmieneh AB, Workie HM, Yadav R, Yadegar A, Yadollahpour A, Yaseri M, Yazdi-Feyzabadi V, Yeshaneh A, Yimam MA, Yimer EM, Yisma E, Yonemoto N, Younis MZ, Yousefi B, Yousefifard M, Yu C, Zabeh E, Zadnik V, Moghadam TZ, Zaidi Z, Zamani M, Zandian H, Zangeneh A, Zaki L, Zendehdel K, Zenebe ZM, Zewale TA, Ziapour A, Zodpey S, Murray CJL. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: a systematic analysis for the global burden of disease study[J]. JAMA Oncol, 2019, 5(12): 1749-1768.
[DOI]
|
| [35] |
Block KI, Gyllenhaal C, Lowe L, Amedei A, Amin A, Amin A, Aquilano K, Arbiser J, Arreola A, Arzumanyan A, Ashraf SS, Azmi AS, Benencia F, Bhakta D, Bilsland A, Bishayee A, Blain SW, Block PB, Boosani CS, Carey TE, Carnero A, Carotenuto M, Casey SC, Chakrabarti M, Chaturvedi R, Chen GZ, Chen H, Chen S, Chen YC, Choi BK, Ciriolo MR, Coley HM, Collins AR, Connell M, Crawford S, Curran CS, Dabrosin C, Damia G, Dasgupta S, Deberardinis RJ, Decker WK, Dhawan P, Diehl AME, Dong JT, Dou QP, Drew JE, Elkord E, El-Rayes B, Feitelson MA, Felsher DW, Ferguson LR, Fimognari C, Firestone GL, Frezza C, Fujii H, Fuster MM, Generali D, Georgakilas AG, Gieseler F, Gilbertson M, Green MF, Grue B, Guha G, Halicka D, Helferich WG, Heneberg P, Hentosh P, Hirschey MD, Hofseth LJ, Holcombe RF, Honoki K, Hsu HY, Huang GS, Jensen LD, Jiang WG, Jones LW, Karpowicz PA, Keith WN, Kerkar SP, Khan GN, Khatami M, Ko YH, Kucuk O, Kulathinal RJ, Kumar NB, Kwon BS, Le A, Lea MA, Lee HY, Lichtor T, Lin LT, Locasale JW, Lokeshwar BL, Longo VD, Lyssiotis CA, Mackenzie KL, Malhotra M, Marino M, Martinez-Chantar ML, Matheu A, Maxwell C, Mcdonnell E, Meeker AK, Mehrmohamadi M, Mehta K, Michelotti GA, Mohammad RM, Mohammed SI, Morre DJ, Muralidhar V, Muqbil I, Murphy MP, Nagaraju GP, Nahta R, Niccolai E, Nowsheen S, Panis C, Pantano F, Parslow VR, Pawelec G, Pedersen PL, Poore B, Poudyal D, Prakash S, Prince M, Raffaghello L, Rathmell JC, Rathmell WK, Ray SK, Reichrath J, Rezazadeh S, Ribatti D, Ricciardiello L, Robey RB, Rodier F, Rupasinghe HPV, Russo GL, Ryan EP, Samadi AK, Sanchez-Garcia I, Sanders AJ, Santini D, Sarkar M, Sasada T, Saxena NK, Shackelford RE, Shantha Kumara HMC, Sharma D, Shin DM, Sidransky D, Siegelin MD, Signori E, Singh N, Sivanand S, Sliva D, Smythe C, Spagnuolo C, Stafforini DM, Stagg J, Subbarayan PR, Sundin T, Talib WH, Thompson SK, Tran PT, Ungefroren H, Vander Heiden MG, Venkateswaran V, Vinay DS, Vlachostergios PJ, Wang Z, Wellen KE, Whelan RL, Yang ES, Yang H, Yang X, Yaswen P, Yedjou C, Yin X, Zhu J, Zollo M. Designing a broad-spectrum integrative approach for cancer prevention and treatment[J]. Semin Cancer Biol, 2015, 35(Suppl): S276-S304.
[DOI]
|
| [36] |
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012[J]. Int J Cancer, 2015, 136(5): E359-E386.
[DOI]
|
| [37] |
Magnusson RS, Mcgrady B, Gostin L, Patterson D, Abou Taleb H. Legal capacities required for prevention and control of noncommunicable diseases[J]. Bull World Health Organ, 2019, 97(2): 108-117.
[DOI]
|
| [38] |
Dickinson A, Xu M, Silén S, Wang Y, Fu Y, Sadeghi M, Toppinen M, Carpén T, Hedman K, Mäkitie A, Söderlund-Venermo M. Newly detected DNA viruses in juvenile nasopharyngeal angiofibroma (JNA) and oral and oropharyngeal squamous cell carcinoma (OSCC/OPSCC)[J]. Eur Arch Otorhinolaryngol, 2019, 276(2): 613-617.
[DOI]
|
| [39] |
Rous P. A sarcoma of the fowl transmissible by an agent separable from the tumor cells[J]. J Exp Med, 1911, 13(4): 397-411.
[DOI]
|
| [40] |
Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt's lymphoma[J]. Lancet, 1964, 1(7335): 702-703.
[DOI]
|
| [41] |
Hinuma Y, Nagata K, Hanaoka M, Nakai M, Matsumoto T, Kinoshita KI, Shirakawa S, Miyoshi I. Adult T-cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera[J]. Proc Natl Acad Sci U S A, 1981, 78(10): 6476-6480.
[DOI]
|
| [42] |
Warren JR, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis[J]. Lancet, 1983, 1(8336): 1273-1275.
[DOI]
|
| [43] |
Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration[J]. Lancet, 1984, 1(8390): 1311-1315.
[DOI]
|
| [44] |
Peek RM Jr, Crabtree JE. Helicobacter infection and gastric neoplasia[J]. J Pathol, 2006, 208(2): 233-248.
[DOI]
|
| [45] |
Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention[J]. Cancer Epidemiol Biomarkers Prev, 2014, 23(5): 700-713.
[DOI]
|
| [46] |
Ansari S, Yamaoka Y. Survival of Helicobacter pylori in gastric acidic territory[J]. Helicobacter, 2017, 22(4): 10.1111/hel.12386. doi: 10.1111/hel.12386.
|
| [47] |
Zhu C, Wang Y, Cai C, Cai Q. Bacterial infection and associated cancers[J]. Adv Exp Med Biol, 2017, 1018: 181-191.
[DOI]
|
| [48] |
Lange D, Oeder C, Waltermann K, Mueller A, Oehme A, Rohrberg R, Marsch W, Fischer M. Bacillary angiomatosis[J]. J Dtsch Dermatol Ges, 2009, 7(9): 767-769.
[DOI]
|
| [49] |
Feng M, Cheng X. Parasite-associated cancers (blood flukes/liver flukes)[J]. Adv Exp Med Biol, 2017, 1018: 193-205.
[DOI]
|
| [50] |
Fried B, Reddy A, Mayer D. Helminths in human carcinogenesis[J]. Cancer Lett, 2011, 305(2): 239-249.
[DOI]
|
| [51] |
Nii-Trebi NI. Emerging and neglected infectious diseases: insights, advances, and challenges[J]. Biomed Res Int, 2017, 2017: 5245021.
[DOI]
|
| [52] |
Maru Y, Hippo Y. Current status of patient-derived ovarian cancer models[J]. Cells, 2019, 8(5): 505.
[DOI]
|
| [53] |
Lewinska A, Adamczyk J, Pajak J, Stoklosa S, Kubis B, Pastuszek P, Slota E, Wnuk M. Curcumin-mediated decrease in the expression of nucleolar organizer regions in cervical cancer (HeLa) cells[J]. Mutat Res Genet Toxicol Environ Mutagen, 2014, 771: 43-52.
[DOI]
|
| [54] |
Yu J, Qin B, Moyer AM, Sinnwell JP, Thompson KJ, Copland JA 3rd, Marlow LA, Miller JL, Yin P, Gao B, Minter-Dykhouse K, Tang X, Mclaughlin SA, Moreno-Aspitia A, Schweitzer A, Lu Y, Hubbard J, Northfelt DW, Gray RJ, Hunt K, Conners AL, Suman VJ, Kalari KR, Ingle JN, Lou Z, Visscher DW, Weinshilboum R, Boughey JC, Goetz MP, Wang L. Establishing and characterizing patient-derived xenografts using pre-chemotherapy percutaneous biopsy and post-chemotherapy surgical samples from a prospective neoadjuvant breast cancer study[J]. Breast Cancer Res, 2017, 19(1): 130.
[DOI]
|
| [55] |
Mullenders J, de Jongh E, Brousali A, Roosen M, Blom JPA, Begthel H, Korving J, Jonges T, Kranenburg O, Meijer R, Clevers HC. Mouse and human urothelial cancer organoids: a tool for bladder cancer research[J]. Proc Natl Acad Sci U S A, 2019, 116(10): 4567-4574.
[DOI]
|
| [56] |
王楚, 高建军, 华国强. 肠道类器官在精准医学中的应用[J]. 中国科学(生命科学), 2017, 47(2): 171-179. [CNKI]
|
| [57] |
Williamson IA, Arnold JW, Samsa LA, Gaynor L, Disalvo M, Cocchiaro JL, Carroll I, Azcarate-Peril MA, Rawls JF, Allbritton NL, Magness ST. A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology[J]. Cell Mol Gastroenterol Hepatol, 2018, 6(3): 301-319.
[DOI]
|
| [58] |
Bartfeld S, Clevers H. Organoids as model for infectious diseases: culture of human and murine stomach organoids and microinjection of Helicobacter pylori[J]. J Vis Exp, 2015(105): 53359.
[DOI]
|
| [59] |
Jiao Y, Zhang YG, Lin Z, Lu R, Xia Y, Meng C, Pan Z, Xu X, Jiao X, Sun J. Salmonella enteritidis effector AvrA suppresses autophagy by reducing beclin-1 protein[J]. Front Immunol, 2020, 11: 686.
[DOI]
|
| [60] |
Saxena K, Blutt SE, Ettayebi K, Zeng XL, Broughman JR, Crawford SE, Karandikar UC, Sastri NP, Conner ME, Opekun AR, Graham DY, Qureshi W, Sherman V, Foulke-Abel J, In J, Kovbasnjuk O, Zachos NC, Donowitz M, Estes MK. Human intestinal enteroids: a new model to study human rotavirus infection, host restriction, and pathophysiology[J]. J Virol, 2015, 90(1): 43-56.
[DOI]
|
| [61] |
Drummond CG, Bolock AM, Ma C, Luke CJ, Good M, Coyne CB. Enteroviruses infect human enteroids and induce antiviral signaling in a cell lineage-specific manner[J]. Proc Natl Acad Sci U S A, 2017, 114(7): 1672-1677.
[DOI]
|
| [62] |
Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng X-L, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK. Replication of human noroviruses in stem cell-derived human enteroids[J]. Science, 2016, 353(6306): 1387-1393.
[DOI]
|
| [63] |
Zhou J, Li C, Zhao G, Chu H, Wang D, Yan HH, Poon VK, Wen L, Wong BH, Zhao X, Chiu MC, Yang D, Wang Y, Au-Yeung RKH, Chan IH, Sun S, Chan JF, To KK, Memish ZA, Corman VM, Drosten C, Hung IF, Zhou Y, Leung SY, Yuen KY. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus[J]. Sci Adv, 2017, 3(11): eaao4966.
[DOI]
|
| [64] |
Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, Ravelli RBG, Paul van Schayck J, Mykytyn AZ, Duimel HQ, van Donselaar E, Riesebosch S, Kuijpers HJH, Schippers D, van de Wetering WJ, de Graaf M, Koopmans M, Cuppen E, Peters PJ, Haagmans BL, Clevers H. SARS-CoV-2 productively infects human gut enterocytes[J]. Science, 2020, 369(6499): 50-54.
[DOI]
|
| [65] |
Zhou J, Li C, Sachs N, Chiu MC, Wong BH, Chu H, Poon VK, Wang D, Zhao X, Wen L, Song W, Yuan S, Wong KK, Chan JF, To KK, Chen H, Clevers H, Yuen KY. Differentiated human airway organoids to assess infectivity of emerging influenza virus[J]. Proc Natl Acad Sci U S A, 2018, 115(26): 6822-6827.
[DOI]
|
| [66] |
Porotto M, Ferren M, Chen YW, Siu Y, Makhsous N, Rima B, Briese T, Greninger AL, Snoeck HW, Moscona A. Authentic modeling of human respiratory virus infection in human pluripotent stem cell-derived lung organoids[J]. mBio, 2019, 10(3): e00723-19.
[DOI]
|
| [67] |
Garcez PP, Loiola EC, Madeiro Da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. Zika virus impairs growth in human neurospheres and brain organoids[J]. Science, 2016, 352(6287): 816-818.
[DOI]
|
| [68] |
Zhang B, He Y, Xu Y, Mo F, Mi T, Shen QS, Li C, Li Y, Liu J, Wu Y, Chen G, Zhu W, Qin C, Hu B, Zhou G. Differential antiviral immunity to Japanese encephalitis virus in developing cortical organoids[J]. Cell Death Dis, 2018, 9(7): 719.
[DOI]
|
| [69] |
Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, Haft A, Vries RG, Grompe M, Clevers H. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration[J]. Nature, 2013, 494(7436): 247-250.
[DOI]
|
| [70] |
Zhao B, Ni C, Gao R, Wang Y, Yang L, Wei J, Lv T, Liang J, Zhang Q, Xu W, Xie Y, Wang X, Yuan Z, Liang J, Zhang R, Lin X. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids[J]. Protein Cell, 2020, 11(10): 771-775.
[DOI]
|
| [71] |
Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, Leopoldi A, Garreta E, Hurtado Del Pozo C, Prosper F, Romero JP, Wirnsberger G, Zhang H, Slutsky AS, Conder R, Montserrat N, Mirazimi A, Penninger JM. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2[J]. Cell, 2020, 181(4): 905-913.
[DOI]
|
| [72] |
Nikolić MZ, Caritg O, Jeng Q, Johnson JA, Sun D, Howell KJ, Brady JL, Laresgoiti U, Allen G, Butler R, Zilbauer M, Giangreco A, Rawlins EL. Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids[J]. Elife, 2017, 6: e26575.
[DOI]
|
| [73] |
Richards DJ, Coyle RC, Tan Y, Jia J, Wong K, Toomer K, Menick DR, Mei Y. Inspiration from heart development: biomimetic development of functional human cardiac organoids[J]. Biomaterials, 2017, 142: 112-123.
[DOI]
|
| [74] |
Wörsdörfer P, I T, Asahina I, Sumita Y, Ergün S. Do not keep it simple: recent advances in the generation of complex organoids[J]. J Neural Transm (Vienna), 2020, 127(11): 1569-1577.
[DOI]
|
| [75] |
Ashok A, Choudhury D, Fang Y, Hunziker W. Towards manufacturing of human organoids[J]. Biotechnol Adv, 2020, 39: 107460.
[DOI]
|
 2021, Vol. 16
2021, Vol. 16


