2019年末,一场由新型冠状病毒引起的肺炎疫情暴发。世界卫生组织(World Health Organization,WHO)将引起该肺炎疫情的病原体命名为2019新型冠状病毒(2019 novel coronavirus,2019-nCoV);2020年1月30日,WHO宣布此次疫情为“国际公共卫生紧急事件”(public health emergency of international concern,PHEIC);2020年2月11日,WHO将2019-nCOV感染所致的肺炎定名为2019冠状病毒病(coronavirus disease 2019,COVID-19)①,而国际病毒分类委员会(International Committee on Taxonomy of Viruses,ICTV)冠状病毒研究小组(Coronaviridae Study Group,CSG)将该病毒命名为严重急性呼吸综合征冠状病毒2型(severe acute respiratory syndrome coronavirus 2,SARS-CoV-2)[1]。多位在中国工作的病毒学专家认为将该病毒命名为SARS-CoV-2容易给公众造成新型冠状病毒肺炎就是SARS的不正确印象,因此而采用不正确的防护措施,所以他们也曾建议将其改为“人类冠状病毒2019(human coronavirus 2019,HCoV-19)” [2-3],与WHO确定的疾病名(COVID-19)保持一致。
① 信息来源于https://www.who.int/emergencies/diseases/novel-coronavirus-2019/interactive-timeline#event-45。
疫苗和药物是防治疫情的重要手段。WHO数据显示,截至2021年11月16日,全球共有194个COVID-19疫苗进入临床前实验阶段,132个进入临床试验阶段,其中28个进入临床Ⅲ期,10个进入临床Ⅳ期②。但目前研发的疫苗均是针对现在流行的SARS-CoV-2设计的,对未来新发与再现的高致病性冠状病毒可能无效。此外,如果SARS-CoV-2在未来发生重大突变,现在有效的疫苗在将来可能变得无效。针对SARS-CoV、中东呼吸综合征冠状病毒(Middle East respiratory syndrome, MERS-CoV)和SARS-CoV-2感染,目前尚无获得批准的特异性抗冠状病毒药物。由于疫苗和新药的研发往往滞后于疫情发展,亟须研发安全、广谱、高效的冠状病毒疫苗和药物,以应对未来新发与再现冠状病毒相关疾病导致的疫情。本文主要介绍广谱冠状病毒疫苗和抗冠状病毒多肽的研究进展。
② 信息来源于https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines。
1 概述冠状病毒属于套式病毒目冠状病毒科,是一类有包膜的正链单链RNA病毒,分为α、β、γ、δ 4个属[4],而在β属中又分为A、B、C、D 4个亚型。目前,共发现7种可以感染人类的冠状病毒,包括α属的HCoV-229E和HCoV-NL63以及β属的HCoV-OC43、HCoV-HKU1、SARS-CoV、MERS-CoV和SARS-CoV-2。其中HCoV-229E、HCoV-NL63和HCoV-OC43感染人后主要引起自限性呼吸道症状[5],但也会在儿童、老人及免疫力低下的患者中引起严重的疾病,甚至威胁生命[6-8];SARS-CoV、MERS-CoV以及SARS-CoV-2感染则导致严重呼吸系统症状及全身症状[9]。
冠状病毒的包膜表面有3个重要的结构蛋白,即刺突蛋白(spike,S蛋白)、包膜蛋白(envelope,E蛋白)和膜蛋白(membrane,M蛋白)。S蛋白包含S1和S2两个亚单位,S2亚单位含有HR1和HR2两个7肽重复域(heptad repeat),它们在病毒感染细胞的过程中发挥着重要作用[10]。冠状病毒可通过表面膜融合和内吞膜融合两种方式感染细胞。冠状病毒的S蛋白属于Ⅰ型膜融合蛋白[11]。在病毒感染细胞的过程中,如果细胞表面有跨膜丝氨酸蛋白酶(TMPRSS2),S蛋白就能在其作用下被切割成S1和S2两个亚单位。S1亚单位含有受体结合域(receptor binding domain,RBD),通过RBD与靶细胞膜上的病毒受体结合,随后S2亚单位发生构象变化,其N端的融合多肽(fusion polypeptide,FP)插入靶细胞膜内。HR1与HR2通过HR1区域中“e”和“g”位置的残基与HR2区域中“a”和“d”位置的残基相互作用形成6-HB核心结构,缩短病毒膜与细胞膜的距离,导致膜融合的发生[12],病毒的基因物质通过膜融合孔注入细胞内进行复制。在细胞表面没有TMPRSS2的情况下,病毒也可以通过内吞的形式进入细胞,在内吞泡中的酸性环境以及组织蛋白酶(cathepsin)的作用下,病毒膜与内吞泡膜融合,把病毒的基因物质释放到细胞内(图 1)。
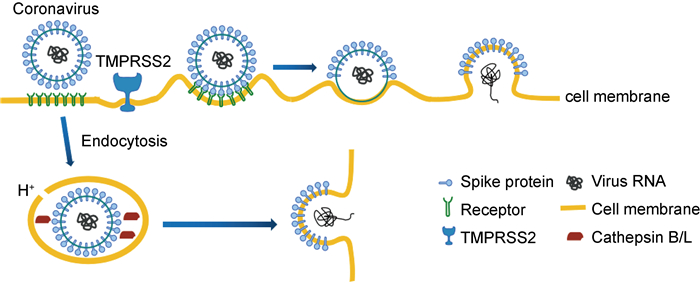
|
| 图 1 冠状病毒的膜融合过程 Fig. 1 Membrane fusion process of coronavirus |
S1亚单位含有的RBD负责介导病毒与宿主细胞表面受体的结合,并含有S蛋白中主要的中和抗体位点,是研发疫苗和中和抗体的重要靶点。S2亚单位含有的HR1和HR2在病毒感染细胞的过程中通过相互作用形成6螺旋(6-HB)核心结构,将病毒膜和靶细胞膜拉近,导致膜融合的发生,病毒基因通过膜融合孔进入宿主细胞内进行复制。HR1和HR2区的序列在不同冠状病毒中相对保守,是设计广谱冠状病毒融合抑制剂的理想靶点。
2 广谱冠状病毒疫苗 2.1 靶抗原的选择冠状病毒的疫苗抗原主要分为病毒颗粒、S蛋白、RBD 3类。研究表明,SARS-CoV-2与SARS-CoV以及Bat-SARSr-CoV(ZC45和ZXC21)的全基因组相似度分别为79%和88%,S1亚单位和S2亚单位基因相似度分别为68%和93%[13]。SARS-CoV-2与SARS-CoV S蛋白的基因相似度为76%~78%,而RBD区域的基因相似度为73%~76%[14]。
大量关于SARS-CoV疫苗的研究显示,特异性体液免疫应答和细胞免疫应答均具有保护作用。已证明靶向SARS-CoV S蛋白的抗体可以保护小鼠免受SARS-CoV的感染[15-17];Liu等[18]的研究显示,携带有SARS-CoV S蛋白的假病毒疫苗免疫小鼠后,诱导产生的血清抗体具有抑制SARS-CoV、SARS相关冠状病毒(SARSr-CoV)如WIV1和Rs3367假病毒感染的能力;基于SARS-CoV RBD设计研发的疫苗免疫小鼠得到的血清抗体,也对SARS-CoV-2假病毒感染具有一定的交叉中和活性,而一些靶向SARS-CoV RBD的中和抗体,如7B11[19]、S309[20]也被证明具有交叉中和抑制SARS-CoV-2感染的能力,这提示基于SARS-CoV RBD研发的疫苗具有成为广谱抗β属冠状病毒疫苗的可能性[21]。
Ahmed等[22]通过分析SARS-CoV来源的115个T细胞表位和298个B细胞表位发现,SARS-CoV-2(序列截至2020年2月21日)存在27个(23%)与SARS-CoV相同的T细胞表位,49个(16%)与SARS-CoV相同的B细胞表位,其中分别有41%和47%的表位位于S蛋白上,这表明相关疫苗有可能在二者间引起T细胞或B细胞反应。此外,他们通过主要组织相容性复合体(major histocompatibility complex, MHC)结合试验发现,SARS-CoV-2与SARS-CoV间存在229个相似的T细胞表位,其中约82%为MHC-I类限制性表位;66个表位来源于S蛋白;且在来源于S蛋白的表位中,3个表位(GYQPYRVVVL、QPYRVVVLSF和PYRVVVLSF)能够完全定位到SARS-CoV的受体结合基序(receptor binding motif,RBM)上。Kiyotani等[23]通过生物信息学对SARS-CoV-2的T细胞表位进行预测也发现,在对人类白细胞抗原(human leukocyte antigen,HLA)具有高亲和力的表位中,约38.8%的HLA-1型表位保守存在于SARS-CoV的3种毒株BJ01、GZ02和Tor2中,约1.8%的表位与MERS-CoV中相关表位具有100%的序列相似性;而在HLA-2型表位中,约29.9%的表位与SARS-CoV的3种毒株相关表位具有100%的序列相似性,其中约2.4%的表位在MERS-CoV中保守。所有这些保守的HLA-1型和HLA-2型表位均主要位于ORFab和S蛋白上[23]。此外,在SARS-CoV和SARS-CoV-2的RBD上与血管紧张素转换酶2(angiotensin-converting enzyme 2, ACE2)相互作用的14个共有氨基酸位置中,二者具有8个相同的氨基酸,5个位置的氨基酸尽管存在不同的侧链,但具有相似的生化特性[24];虽然SASR-CoV的RBD与MERS-CoV的RBD识别不同的受体,在氨基酸序列上具有较低的序列同源性,但在结构上却有高度相似的核心亚结构域[25]。这些研究提示,SARS-CoV和SARS-CoV-2的S蛋白以及RBD具有作为广谱抗β冠状病毒疫苗抗原的可能性。
针对SARS-CoV和MERS-CoV的疫苗研究发现,用灭活的SARS-CoV或MERS-CoV颗粒免疫小鼠,攻毒后会导致小鼠肺部嗜酸性粒细胞浸润及产生偏向Th2型免疫应答的病理损伤[26-28];用表达SARS-CoV S蛋白的重组疫苗免疫雪貂后,虽然动物体内能够产生高效中和抗体,但用SARS-CoV攻毒会导致转氨酶升高,出现严重的肝损伤[29];使用基于S蛋白的SARS-CoV疫苗免疫中国猕猴后,实验动物体内产生抗体依赖性增强(antibody-dependent enhancement,ADE)效应[30]。研究发现,SARS-CoV S蛋白中位于RBD区域(S318-510)以外的表位S597-603可以诱导实验动物产生具有ADE效应的非中和抗体[31],而靶向SARS-CoV S蛋白的一些中和抗体也能通过ADE效应增强病毒感染[32]。所以,非中和抗体和一些中和抗体均具有产生ADE效应的可能性[33],且可能与病毒种类[34]、抗体亚型[35]、抗体浓度[36]、抗体结构[37]、感染剂量以及疫苗接种引起的免疫应答类型等[30]有关。许多病毒感染过程中均可观察到ADE现象。ADE发生的确切分子机制目前尚不十分清楚,研究认为,可能是抗体与病毒结合后,再与细胞表面IgG Fc受体(FcR)结合,介导病毒进入细胞,从而导致感染增强[33]。此外,对埃博拉病毒的致病机制研究发现,病毒与不同血清型的抗体结合可激活经典的补体途径,促进病毒感染[38]。
当然,也许SARS-CoV-2与SARS-CoV的免疫原性大不相同。恒河猴攻毒实验结果显示,接种6 μg的SARS-CoV-2灭活疫苗(PiCoVacc)可以起到保护作用,且未出现ADE效应[39]。Ⅰ期临床试验证明,基于S蛋白的新型5型腺病毒载体COVID-19(Ad5-nCoV)疫苗仅使用单剂量即可在14 d内产生病毒特异性抗体和T细胞免疫反应,且未出现严重不良反应[40]。
RBD不仅是识别受体的重要结构域,也是病毒感染机体后中和抗体识别的主要靶点[41]。基于RBD的疫苗不易诱导具有ADE效应的非中和抗体[42-45],且不易在免疫动物体内产生Th2型免疫应答损伤及肺部嗜酸性粒细胞浸润[46]。而与靶向S1亚单位(不包括RBD结合域)的单克隆抗体相比,RBD特异性的单克隆抗体具有更强的中和能力[47]。此外,CD8+T细胞反应在病毒清除、避免产生嗜酸性细胞免疫病理损伤和保护机体免受病毒感染方面发挥了重要作用[34, 48-49],而SARS-CoV、MERS-CoV和SARS-CoV-2等冠状病毒的S蛋白上也存在一些保守的T细胞表位,这些表位部分位于RBD[22-23]。因此,RBD具有较高的安全性和免疫原性,是研发安全、有效、广谱冠状病毒疫苗的最佳抗原选择。
2.2 抗原优化虽然RBD含有S蛋白中主要的中和抗体位点,但也有一些具有免疫优势的非中和位点[50-51],可刺激机体产生无用或有害的非中和免疫反应。RBD的中和免疫原性可使用“中和免疫原性指数(neutralizing immunogenicity index,NII)”和“广谱中和免疫原性指数(broad neutralizing immunogenicity index,BNII)”进行评估[52],并采用“摘帽”(去糖基化)和“戴帽”(糖基化)的策略对RBD进行优化。如果RBD中某些中和抗体位点被糖基化“帽子”掩盖了,可将其糖基化“帽子”去除,而那些非中和抗体位点可用糖基化“帽子”遮盖。这样不但能提高RBD的免疫原性,也可提高其广谱性。RBD中部分保守区域也可诱导机体产生一些广谱中和抗体,但由于具有免疫优势的非中和抗体显性表位存在,转移了宿主的免疫反应,从而影响了广谱中和抗体的产生[51, 53]。此外,当去除S蛋白的其他部分而仅保留RBD时,重组RBD疫苗会暴露出大量被隐藏的可能包含具有免疫优势的非中和位点的表位,从而影响广谱中和抗体的产生[52]。如果将这些非中和抗体显性表位“戴帽”遮盖后,比较保守的中和抗体位点就占据了优势,可能产生更多的广谱中和抗体。
2.3 佐剂选择使用佐剂可有效增强抗原的免疫原性,降低抗原的使用量。铝佐剂、MF59佐剂、AS03以及Toll样受体激动剂是常用的疫苗佐剂,通常以肌内或皮内注射的方式进行免疫。与皮下免疫相比,基于RBD的疫苗通过滴鼻免疫能够在小鼠肺部引起更强的全身性细胞免疫反应和更高的局部黏膜免疫应答[54]。Wang等[55]基于干扰素基因刺激蛋白(stimulator of interferon genes, STING)激动剂环鸟苷酸-腺苷酸(cyclin GMP-AMP,cGAMP),设计了一种通过鼻腔免疫的方式来模拟流感病毒感染肺部的仿生纳米颗粒(PS-GAMP)。这种方法可有效激活肺泡巨噬细胞和上皮细胞,促进疫苗产生高效的体液免疫应答和细胞免疫应答,从而能够使H1N1季节性流感疫苗成为预防多种异型流感病毒感染的广谱流感疫苗[55]。因此,可结合使用PS-GAMP仿生纳米佐剂与基于SARS-CoV-2的RBD疫苗,通过鼻腔免疫诱导机体产生高效、广谱抗冠状病毒的中和抗体和T细胞免疫应答,增强疫苗的免疫原性和广谱性。
3 广谱抗冠状病毒多肽 3.1 冠状病毒感染细胞的过程在SARS-CoV和MERS-CoV经表面膜融合形成的6-HB核心结构中,HR1与HR2之间许多区域具有很强的相互作用,称为HR1核心区和HR2核心区。通过序列比对发现,SARS-CoV-2与SARS-CoV在HR1与HR2区域分别具有92.6%和100%的序列相似性,但在HR1核心区的21个氨基酸中存在8个氨基酸的突变,这可能影响HR1与HR2的相互作用模式,从而影响6-HB束的形成[56]。为探究SARS-CoV-2的膜融合机制,Xia等[56]合成了SARS-CoV-2的HR1和HR2两条多肽,即SARS-CoV-2-HR1P和SARS-CoV-2-HR2P,发现二者能够在体外形成6-HB结构,证明SARS-CoV-2与SARS-CoV、MERS-CoV具有相同的膜融合机制。晶体结构解析发现,HR1核心区域的8个氨基酸突变不但不影响6-HB的形成,反而能够加强HR1与HR2的相互作用,进一步稳定6-HB结构,赋予SARS-CoV-2比SARS-CoV更强的膜融合能力[57]。
3.2 多肽类冠状病毒抑制剂阻断病毒与细胞膜融合的发生是抑制冠状病毒感染细胞的重要手段。S蛋白S2亚单位的HR1和HR2区域在各冠状病毒中高度保守,并且二者在膜融合过程中相互作用形成6-HB的机制也相对保守[58]。此外,在膜融合过程中HR1区域只是瞬间暴露,很难诱导产生药物耐受突变,因此HR1区域是广谱冠状病毒融合抑制剂开发的最佳靶点。由HR1与HR2所形成融合中间态的空间比较小,只允许相对分子质量(MW)<7×104的小分子蛋白质或多肽进入。而抗体分子太大(MW约1.5×105),难以进入该区域发挥作用;小分子药物又太小,不足以阻止HR1和HR2的相互作用,因此多肽是针对该靶点最适合研发的融合抑制剂。
已有研究报道,包膜病毒如人类免疫缺陷病毒(HIV)[59-60]、呼吸道合胞病毒(RSV)[61]、埃博拉病毒[62]、副黏病毒(SV5)[63]、尼帕病毒[64]以及鼠肝炎病毒(MHV)[65]的Ⅰ型膜融合蛋白中来源于HR2区域的多肽,能够通过与相关病毒S蛋白HR1区域结合形成6-HB,从而有效抑制病毒S蛋白介导的膜融合。既往研究也表明,来源于SARS-CoV和MERS-CoV S蛋白HR2区域的衍生多肽,可以和病毒的HR2区域竞争性地与HR1结合,抑制病毒形成6-HB结构,从而阻止病毒融合和进入宿主细胞[58, 66]。体外研究表明,来自SARS-CoV-2的HR2衍生肽也能有效抑制SARS-CoV-2假病毒的感染[56]。尽管衍生于SARS-CoV、MERS-CoV和SARS-CoV-2的HR2区的多肽能特异性地抑制其相应病毒的膜融合,但这些多肽均不能抑制异源冠状病毒的感染。
3.3 多肽类广谱冠状病毒抑制剂 3.3.1 EK1为进一步提升HR2衍生肽对冠状病毒的广谱抑制活性,Xia等[56]通过序列比对定位α属冠状病毒(HCoV-NL63、HCoV-229E)和β属冠状病毒(SARS-CoV、MERS-CoV和HCoV-OC43)S蛋白中HR1和HR2的保守区域,合成其衍生肽NL63-HR1P、NL63-HR2P、229E-HR1P、229E-HR2P、SARS-HR1P、SARS-HR2P、MERS-HR1P、MERS-HR2P、OC43-HR1P以及OC43-HR2P。使用不同冠状病毒S蛋白介导的细胞融合实验检测衍生肽对膜融合的抑制活性,最终筛选出对多种冠状病毒与细胞膜融合具有抑制活性的衍生肽OC43-HR2P。通过在i和i+3及i和i+4位置引入E-K盐桥,并在预期不会参与HR1结合的位置引入4Q、14Y、32D和36L突变,得到EK0、EK1、EK2和EK3这4条优化多肽,从而筛选出具有广谱冠状病毒融合抑制活性且溶解性良好的多肽EK1。结构生物学研究显示,EK1能与上述所有HCoV包括SARS-CoV-2的S蛋白HR1区结合,其相互作用的位点非常保守[56]。此外,EK1在HCoV-OC43及MERS-CoV感染的小鼠模型中表现出较高的安全性、有效性及较低的免疫原性,预示着其在临床上具有应用潜力[11]。
3.3.2 EK1C4许多研究表明,通过对多肽进行脂化修饰可以有效提高多肽类病毒融合/进入抑制剂的抗病毒活性[67-68]。因此,Xia等[56]以4个重复聚乙二醇(polyethylene glycol, PEG)为间隔子,在EK1的C端分别利用棕榈酸和胆固醇修饰,合成了EK1P和EK1C两条多肽,二者对SARS-CoV-2的抑制活性分别为69.2 nmol/L和48.1 nmol/L,表明与棕榈酸修饰相比,胆固醇修饰的脂化策略更优。
基于EK1C的分子基础,通过在EK1和胆固醇之间构建不同长度的GSG连接子和PEG连接子,合成了7个EK1C的衍生肽(图 2)。其中EK1C4表现出最高的抗SARS-CoV-2活性,其对SARS-CoV-2的S蛋白介导的细胞融合及假病毒感染的半数抑制浓度(50 % inhibition concentration,IC50)分别为1.3 nmol/L和15.8 nmol/L,抑制能力分别是原始多肽EK1的241倍和149倍。此外,EK1C4对包括SARS-CoV、MERS-CoV、HCoV-OC43、HCoV-NL63和HCoV-229E在内的HCoV也具有优于EK1的抑制活性,并且能够抑制SARSr-WIV1、SARSr-Rs3367两种蝙蝠冠状病毒的感染[57]。上述研究表明EK1C4具有广谱高效的抗冠状病毒活性(图 3),将来有望以鼻腔喷雾或者吸入剂的形式预防和治疗各类冠状病毒感染。

|
| 图 2 EK1脂肽设计 Fig. 2 Design of EK1-lipopeptides |
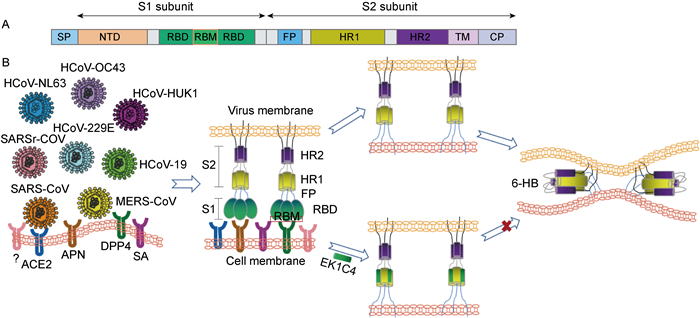
|
| 图 3 冠状病毒S蛋白结构(A)及EK1C4的广谱抗冠状病毒机制(B) Fig. 3 Structure of coronavirus S protein (A) and the mechanism of broad-spectrum anti-coronavirus agent-EK1C4 (B) |
从2003年的SARS-CoV到2012年的MERS-CoV,再到2019年的SARS-CoV-2,冠状病毒疫情的不断发生给人类健康造成巨大的威胁,但截至目前,全球仍无能够有效预防冠状病毒感染的广谱疫苗或特效药。而在未来,也无法保证SARS-CoV、MERS-CoV以及SARS-CoV-2等人类高致病性冠状病毒导致的疫情不会卷土重来,因此研发广谱冠状病毒疫苗和药物是防治冠状病毒相关疾病的重要手段。
在广谱疫苗的研发上,首先要寻找具有广谱免疫原性的抗原,兼顾其安全性及有效性,而RBD是一个理想的位点。不同冠状病毒RBD上,中和抗体与病毒结合的重要位点即抗体结合域(receptor binding motif,RBM)具有很高的变异性[69],难以在不同冠状病毒之间诱导产生针对该区域的广谱中和抗体,但也有研究表明,许多中和抗体识别的目标位点位于RBM之外[70-72],并能通过这种识别改变RBD构象,从而影响RBD与受体结合[72]。因此在广谱疫苗的设计上,采用NII和BNII两个指数对疫苗的有效性和广谱性进行评估,可实现对RBD的抗原位点持续性优化,从而设计出广谱、高效的冠状病毒疫苗。
在广谱抗冠状病毒药物研究上,除现在进行临床测试的许多小分子化合物类如RNA依赖的RNA聚合酶(RdRp)抑制剂和主要蛋白酶(Mpro)抑制剂外,多肽类病毒融合/进入抑制剂也是具有开发前景的候选药物。靶向冠状病毒S蛋白HR1区的多肽类融合/进入抑制剂EK1和EK1C4,在体内外实验中都表现出广谱、高效的抗病毒活性,并且在动物体内也具有较高的安全性[70-72]。此外,与抗体药物相比,多肽药物具有成本低、易规模化生产、易储存和运输的特点,亦可通过鼻腔喷雾或雾化吸入的剂型来预防和治疗新型冠状病毒感染,具有用于防控未来新发与再发冠状病毒疫情的前景。
| [1] |
Lauxmann MA, Santucci NE, Autran-Gomez AM. The SARS-CoV-2 coronavirus and the COVID-19 outbreak[J]. Int Braz J Urol, 2020, 46(suppl.1): 6-18.
|
| [2] |
Jiang SB, Shi ZL, Shu YL, Song JD, Gao GF, Tan WJ, Guo DY. A distinct name is needed for the new coronavirus[J]. Lancet, 2020, 395(10228): 949.
[DOI]
|
| [3] |
张冬梅, 姜世勃, 石正丽, 谭文杰, 卢洪洲, 曹彬, 张文宏, 刘叔文, 钟劲, 肖庚富, 谢幼华, 邓凯, 赵金存, 杨子峰, 宋敬东, 陆路, 黄竞荷, 应天雷, 王乔, 施莽, 徐建青, 安静, 鲁凤民, 高福, 舒跃龙, 郭德银. 关于新型冠状病毒命名的思考与建议[J]. 中国科技术语, 2020, 22(2): 5-10. [DOI]
|
| [4] |
Heinze C. A novel mycovirus from Clitocybe odora[J]. Arch Virol, 2012, 157(9): 1831-1834.
[DOI]
|
| [5] |
Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF. Epidemiology, genetic recombination, and pathogenesis of coronaviruses[J]. Trends Microbiol, 2016, 24(6): 490-502.
[DOI]
|
| [6] |
Simon A, Volz S, Hofling K, Kehl A, Tillman R, Muller A, Kupfer B, Eis-Hubinger AM, Lentze MJ, Bode U, Schildgen O. Acute life threatening event (ALTE) in an infant with human coronavirus HCoV-229E infection[J]. Pediatr Pulmonol, 2007, 42(4): 393-396.
[DOI]
|
| [7] |
Mayer K, Nellessen C, Hahn-Ast C, Schumacher M, Pietzonka S, Eis-Hubinger AM, Drosten C, Brossart P, Wolf D. Fatal outcome of human coronavirus NL63 infection despite successful viral elimination by IFN-alpha in a patient with newly diagnosed ALL[J]. Eur J Haematol, 2016, 97(2): 208-210.
[DOI]
|
| [8] |
Morfopoulou S, Brown JR, Davies EG, Anderson G, Virasami A, Qasim W, Chong WK, Hubank M, Plagnol V, Desforges M, Jacques TS, Talbot PJ, Breuer J. Human coronavirus OC43 associated with fatal encephalitis[J]. N Engl J Med, 2016, 375(5): 497-498.
[DOI]
|
| [9] |
de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses[J]. Nat Rev Microbiol, 2016, 14(8): 523-534.
[DOI]
|
| [10] |
Du L, Yang Y, Zhou Y, Lu L, Li F, Jiang S. MERS-CoV spike protein: a key target for antivirals[J]. Expert Opin Ther Targets, 2017, 21(2): 131-143.
[DOI]
|
| [11] |
Xia S, Yan L, Xu W, Agrawal AS, Algaissi A, Tseng CK, Wang Q, Du L, Tan W, Wilson IA, Jiang S, Yang B, Lu L. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike[J]. Sci Adv, 2019, 5(4): eaav4580.
[DOI]
|
| [12] |
Xia S, Liu Q, Wang Q, Sun Z, Su S, Du L, Ying T, Lu L, Jiang S. Middle East respiratory syndrome coronavirus (MERS-CoV) entry inhibitors targeting spike protein[J]. Virus Res, 2014, 194: 200-210.
[DOI]
|
| [13] |
Lu RJ, Zhao X, Li J, Niu PH, Yang B, Wu HL, Wang WL, Song H, Huang BY, Zhu N, Bi YH, Ma XJ, Zhan FX, Wang L, Hu T, Zhou H, Hu ZH, Zhou WM, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan JY, Xie ZH, Ma JM, Liu WJ, Wang DY, Xu WB, Holmes EC, Gao GF, Wu GZ, Chen WJ, Shi WF, Tan WJ. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding[J]. Lancet, 2020, 395(10224): 565-574.
[DOI]
|
| [14] |
Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus[J]. J Virol, 2020, 94(7): e00127-00120.
[DOI]
|
| [15] |
Yang ZY, Kong WP, Huang Y, Roberts A, Murphy BR, Subbarao K, Nabel GJ. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice[J]. Nature, 2004, 428(6982): 561-564.
[DOI]
|
| [16] |
Deming D, Sheahan T, Heise M, Yount B, Davis N, Sims A, Suthar M, Harkema J, Whitmore A, Pickles R, West A, Donaldson E, Curtis K, Johnston R, Baric R. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants (vol 3, pg 2359, 2006)[J]. PLoS Med, 2006, 3(12): e525.
[DOI]
|
| [17] |
Graham RL, Becker MM, Eckerle LD, Bolles M, Denison MR, Baric RS. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease[J]. Nat Med, 2012, 18(12): 1820-1826.
[DOI]
|
| [18] |
Liu Z, Xia S, Wang X, Lan Q, Xu W, Wang Q, Jiang S, Lu L. Inefficiency of sera from mice treated with pseudotyped SARS-CoV to neutralize 2019-nCoV infection[J]. Virol Sin, 2020, 35(3): 340-343.
[DOI]
|
| [19] |
Tai W, Zhang X, He Y, Jiang S, Du L. Identification of SARS-CoV RBD-targeting monoclonal antibodies with cross-reactive or neutralizing activity against SARS-CoV-2[J]. Antiviral Res, 2020, 179: 104820.
[DOI]
|
| [20] |
Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, Peter A, Guarino B, Spreafico R, Cameroni E, Case JB, Chen RE, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Lanzavecchia A, Diamond MS, Fink K, Veesler D, Corti D. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody[J]. Nature, 2020, 583(7815): 290-295.
[DOI]
|
| [21] |
Tai WB, He L, Zhang XJ, Pu J, Voronin D, Jiang SB, Zhou YS, Du LY. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine[J]. Cell Mol Immunol, 2020, 17(6): 613-620.
[DOI]
|
| [22] |
Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies[J]. Viruses, 2020, 12(3): 254.
[DOI]
|
| [23] |
Kiyotani K, Toyoshima Y, Nemoto K, Nakamura Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2[J]. J Hum Genet, 2020, 65(7): 569-575.
[DOI]
|
| [24] |
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor[J]. Nature, 2020, 581(7807): 215-220.
[DOI]
|
| [25] |
Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, Guo D, Fu L, Cui Y, Liu X, Arledge KC, Chen YH, Zhang L, Wang X. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4[J]. Cell Res, 2013, 23(8): 986-993.
[DOI]
|
| [26] |
Tseng CT, Sbrana E, Iwata-Yoshikawa N, Newman PC, Garron T, Atmar RL, Peters CJ, Couch RB. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus[J]. Plos One, 2012, 7(4): e35421.
[DOI]
|
| [27] |
Agrawal AS, Tao XR, Algaissi A, Garron T, Narayanan K, Peng BH, Couch RB, Tseng CTK. Immunization with inactivated Middle East Respiratory Syndrome coronavirus vaccine leads to lung immunopathology on challenge with live virus[J]. Hum Vaccin Immunother, 2016, 12(9): 2351-2356.
[DOI]
|
| [28] |
Deng Y, Lan J, Bao L, Huang B, Ye F, Chen Y, Yao Y, Wang W, Qin C, Tan W. Enhanced protection in mice induced by immunization with inactivated whole viruses compare to spike protein of middle east respiratory syndrome coronavirus[J]. Emerg Microbes Infect, 2018, 7(1): 60.
[DOI]
|
| [29] |
Weingartl H, Czub M, Czub S, Neufeld J, Marszal P, Gren J, Smith G, Jones S, Proulx R, Deschambault Y, Grudeski E, Andonov A, He RT, Li Y, Copps J, Grolla A, Dick D, Berry J, Ganske S, Manning L, Cao JX. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets[J]. J Virol, 2004, 78(22): 12672-12676.
[DOI]
|
| [30] |
Liu L, Wei Q, Lin Q, Fang J, Wang H, Kwok H, Tang H, Nishiura K, Peng J, Tan Z, Wu T, Cheung KW, Chan KH, Alvarez X, Qin C, Lackner A, Perlman S, Yuen KY, Chen Z. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection[J]. JCI Insight, 2019, 4(4): e123158.
[DOI]
|
| [31] |
Wang QD, Zhang LF, Kuwahara K, Li L, Liu ZJ, Li TS, Zhu H, Liu JN, Xu YF, Xie J, Morioka H, Sakaguchi N, Qjn C, Liu G. Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human primates[J]. ACS Infect Dis, 2016, 2(5): 361-376.
[DOI]
|
| [32] |
Wang SF, Tseng SP, Yen CH, Yang JY, Tsao CH, Shen CW, Chen KH, Liu FT, Liu WT, Chen YMA, Huang JC. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins[J]. Biophys Res Commun, 2014, 451(2): 208-214.
[DOI]
|
| [33] |
Wan Y, Shang J, Sun S, Tai W, Chen J, Geng Q, He L, Chen Y, Wu J, Shi Z, Zhou Y, Du L, Li F. Molecular mechanism for antibody-dependent enhancement of coronavirus entry[J]. J Virol, 2020, 94(5): e02015-e02019.
[DOI]
|
| [34] |
Huisman W, Martina BE, Rimmelzwaan GF, Gruters RA, Osterhaus AD. Vaccine-induced enhancement of viral infections[J]. Vaccine, 2009, 27(4): 505-512.
[DOI]
|
| [35] |
Wang TT, Sewatanon J, Memoli MJ, Wrammert J, Bournazos S, Bhaumik SK, Pinsky BA, Chokephaibulkit K, Onlamoon N, Pattanapanyasat K, Taubenberger JK, Ahmed R, Ravetch JV. IgG antibodies to dengue enhanced for FcγRⅢA binding determine disease severity[J]. Science, 2017, 355(6323): 395-398.
[DOI]
|
| [36] |
Ripoll DR, Wallqvist A, Chaudhury S. Molecular simulations reveal the role of antibody fine specificity and viral maturation state on antibody-dependent enhancement of infection in Dengue virus[J]. Front Cell Infect Microbiol, 2019, 9: 200.
[DOI]
|
| [37] |
Morrone SR, Lok SM. Structural perspectives of antibody-dependent enhancement of infection of dengue virus[J]. Curr Opin Virol, 2019, 36: 1-8.
[DOI]
|
| [38] |
Takada A, Feldmann H, Ksiazek TG, Kawaoka Y. Antibody-dependent enhancement of Ebola virus infection[J]. J Virol, 2003, 77(13): 7539-7544.
[DOI]
|
| [39] |
Gao Q, Bao L, Mao H, Wang L, Xu K, Yang M, Li Y, Zhu L, Wang N, Lv Z, Gao H, Ge X, Kan B, Hu Y, Liu J, Cai F, Jiang D, Yin Y, Qin C, Li J, Gong X, Lou X, Shi W, Wu D, Zhang H, Zhu L, Deng W, Li Y, Lu J, Li C, Wang X, Yin W, Zhang Y, Qin C. Development of an inactivated vaccine candidate for SARS-CoV-2[J]. Science, 2020, 369(6499): 77-81.
[DOI]
|
| [40] |
Zhu FC, Li YH, Guan XH, Hou LH, Wang WJ, Li JX, Wu SP, Wang BS, Wang Z, Wang L, Jia SY, Jiang HD, Wang L, Jiang T, Hu Y, Gou JB, Xu SB, Xu JJ, Wang XW, Wang W, Chen W. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial[J]. Lancet, 2020, 395(10240): 1845-1854.
[DOI]
|
| [41] |
Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV--a target for vaccine and therapeutic development[J]. Nat Rev Microbiol, 2009, 7(3): 226-236.
[DOI]
|
| [42] |
He Y, Li J, Du L, Yan X, Hu G, Zhou Y, Jiang S. Identification and characterization of novel neutralizing epitopes in the receptor-binding domain of SARS-CoV spike protein: revealing the critical antigenic determinants in inactivated SARS-CoV vaccine[J]. Vaccine, 2006, 24(26): 5498-5508.
[DOI]
|
| [43] |
He Y, Zhou Y, Liu S, Kou Z, Li W, Farzan M, Jiang S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine[J]. Biochem Biophys Res Commun, 2004, 324(2): 773-781.
[DOI]
|
| [44] |
He YX, Zhu QY, Liu SW, Zhou YS, Yang BA, Li JM, Jiang SB. Identification of a critical neutralization determinant of severe acute respiratory syndrome (SARS)-associated coronavirus: importance for designing SARS vaccines[J]. Virology, 2005, 334(1): 74-82.
[DOI]
|
| [45] |
He Y, Li J, Li W, Lustigman S, Farzan M, Jiang S. Cross-neutralization of human and palm civet severe acute respiratory syndrome coronaviruses by antibodies targeting the receptor-binding domain of spike protein[J]. J Immunol, 2006, 176(10): 6085-6092.
[DOI]
|
| [46] |
Jiang SB, Bottazzi ME, Du LY, Lustigman S, Tseng CTK, Curti E, Jones K, Zhan B, Hotez PJ. Roadmap to developing a recombinant coronavirus S protein receptor-binding domain vaccine for severe acute respiratory syndrome[J]. Expert Rev Vaccines, 2012, 11(12): 1405-1413.
[DOI]
|
| [47] |
Wang LS, Shi W, Chappell JD, Joyce MG, Zhang Y, Kanekiyo M, Becker MM, van Doremalen N, Fischer R, Wang NS, Corbett KS, Choe M, Mason RD, Van Galen JG, Zhou TQ, Saunders KO, Tatti KM, Haynes LM, Kwong PD, Modjarrad K, Kong WP, McLellan JS, Denison MR, Munster VJ, Mascola JR, Graham BS. Importance of neutralizing monoclonal antibodies targeting multiple antigenic sites on the middle east respiratory syndrome coronavirus spike glycoprotein to avoid neutralization escape[J]. J Virol, 2018, 92(10): e02002-17.
[DOI]
|
| [48] |
Channappanavar R, Fett C, Zhao J, Meyerholz DK, Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection[J]. J Virol, 2014, 88(19): 11034-11044.
[DOI]
|
| [49] |
Honda-Okubo Y, Barnard D, Ong CH, Peng BH, Tseng CTK, Petrovsky N. Severe acute respiratory syndrome-associated coronavirus vaccines formulated with delta inulin adjuvants provide enhanced protection while ameliorating lung eosinophilic immunopathology[J]. J Virol, 2015, 89(6): 2995-3007.
[DOI]
|
| [50] |
Kulp DW, Schief WR. Advances in structure-based vaccine design[J]. Curr Opin Virol, 2013, 3(3): 322-331.
[DOI]
|
| [51] |
Kwong PD, Mascola JR, Nabel GJ. Rational design of vaccines to elicit broadly neutralizing antibodies to HIV-1[J]. Cold Spring Harb Perspect Med, 2011, 1(1): a007278.
[DOI]
|
| [52] |
Du L, Tai W, Yang Y, Zhao G, Zhu Q, Sun S, Liu C, Tao X, Tseng CK, Perlman S, Jiang S, Zhou Y, Li F. Introduction of neutralizing immunogenicity index to the rational design of MERS coronavirus subunit vaccines[J]. Nat Commun, 2016, 7: 13473.
[DOI]
|
| [53] |
Cirelli KM, Crotty S. Germinal center enhancement by extended antigen availability[J]. Curr Opin Immunol, 2017, 47: 64-69.
[DOI]
|
| [54] |
Ma CQ, Li Y, Wang LL, Zhao GY, Tao XR, Tseng CTK, Zhou YS, Du LY, Jiang SB. Intranasal vaccination with recombinant receptor-binding domain of MERS-CoV spike protein induces much stronger local mucosal immune responses than subcutaneous immunization: implication for designing novel mucosal MERS vaccines[J]. Vaccine, 2014, 32(18): 2100-2108.
[DOI]
|
| [55] |
Wang J, Li P, Yu Y, Fu Y, Jiang H, Lu M, Sun Z, Jiang S, Lu L, Wu MX. Pulmonary surfactant-biomimetic nanoparticles potentiate heterosubtypic influenza immunity[J]. Science, 2020, 367(6480): eaau0810.
[DOI]
|
| [56] |
Xia S, Zhu Y, Liu M, Lan Q, Xu W, Wu Y, Ying T, Liu S, Shi Z, Jiang S, Lu L. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein[J]. Cell Mol Immunol, 2020, 17(7): 765-767.
[DOI]
|
| [57] |
Xia S, Liu MQ, Wang C, Xu W, Lan QS, Feng SL, Qi FF, Bao LL, Du LY, Liu SW, Qin C, Sun F, Shi ZL, Zhu Y, Jiang SB, Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion[J]. Cell Res, 2020, 30(4): 343-355.
[DOI]
|
| [58] |
Liu SW, Xiao GF, Chen YB, He YX, Niu JK, Escalante CR, Xiong HB, Farmar J, Debnath AK, Tien P, Jiang SB. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors[J]. Lancet, 2004, 363(9413): 938-947.
[DOI]
|
| [59] |
Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection[J]. Proc Natl Acad Sci U S A, 1994, 91(21): 9770-9774.
[DOI]
|
| [60] |
Jiang SB, Lin K, Strick N, Neurath AR. Hiv-1 inhibition by a peptide[J]. Nature, 1993, 365(6442): 113-113.
[DOI]
|
| [61] |
Lawless-Delmedico MK, Sista P, Sen R, Moore NC, Antczak JB, White JM, Greene RJ, Leanza KC, Matthews TJ, Lambert DM. Heptad-repeat regions of respiratory syncytial virus F1 protein form a six-membered coiled-coil complex[J]. Biochemistry, 2000, 39(38): 11684-11695.
[DOI]
|
| [62] |
Watanabe S, Takada A, Watanabe T, Ito H, Kida H, Kawaoka Y. Functional importance of the coiled-coil of the Ebola virus glycoprotein[J]. J Virol, 2000, 74(21): 10194-10201.
[DOI]
|
| [63] |
Russell CJ, Jardetzky TS, Lamb RA. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion[J]. EMBO J, 2001, 20(15): 4024-4034.
[DOI]
|
| [64] |
Bossart KN, Mungall BA, Crameri G, Wang LF, Eaton BT, Broder CC. Inhibition of Henipavirus fusion and infection by heptad-derived peptides of the Nipah virus fusion glycoprotein[J]. Virol J, 2005, 2: 57.
[DOI]
|
| [65] |
Bosch BJ, van der Zee R, de Haan CAM, Rottier PJM. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex[J]. J Virol, 2003, 77(16): 8801-8811.
[DOI]
|
| [66] |
Lu L, Liu Q, Zhu Y, Chan KH, Qin L, Li Y, Wang Q, Chan JF, Du L, Yu F, Ma C, Ye S, Yuen KY, Zhang R, Jiang S. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor[J]. Nat Commun, 2014, 5: 3067.
[DOI]
|
| [67] |
Mathieu C, Porotto M, Figueira TN, Horvat B, Moscona A. Fusion inhibitory lipopeptides engineered for prophylaxis of Nipah virus in primates[J]. J Infect Dis, 2018, 218(2): 218-227.
[DOI]
|
| [68] |
Chong H, Xue J, Xiong S, Cong Z, Ding X, Zhu Y, Liu Z, Chen T, Feng Y, He L, Guo Y, Wei Q, Zhou Y, Qin C, He Y. A lipopeptide HIV-1/2 fusion inhibitor with highly potent in vitro, ex vivo, and in vivo antiviral activity[J]. J Virol, 2017, 91(11): e00288-17.
[DOI]
|
| [69] |
Freund NT, Roitburd-Berman A, Sui J, Marasco WA, Gershoni JM. Reconstitution of the receptor-binding motif of the SARS coronavirus[J]. Protein Eng Des Sel, 2015, 28(12): 567-575.
[DOI]
|
| [70] |
He YX, Li JJ, Heck S, Lustigman S, Jiang SB. Antigenic and immunogenic characterization of recombinant baculovirus-expressed severe acute respiratory syndrome coronavirus spike protein: implication for vaccine design[J]. J Virol, 2006, 80(12): 5757-5767.
[DOI]
|
| [71] |
Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, Lu L, Jiang S, Yang Z, Wu Y, Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody[J]. Emerg Microbes Infect, 2020, 9(1): 382-385.
[DOI]
|
| [72] |
Chen WH, Hotez PJ, Bottazzi ME. Potential for developing a SARS-CoV receptor-binding domain (RBD) recombinant protein as a heterologous human vaccine against coronavirus infectious disease (COVID)-19[J]. Hum Vaccin Immunother, 2020, 16(6): 1239-1242.
[DOI]
|
 2021, Vol. 16
2021, Vol. 16


