2. 复旦大学附属上海市公共卫生临床中心,上海 201508
2. Shanghai Public Health Clinical Center, Fudan University, Shanghai 201508, China
人类诺如病毒(human norovirus,HuNoV)属于杯状病毒科诺如病毒属,为无包膜的单股正链RNA病毒[1]。其基因组长约7.5 kb,包含3个开放阅读框(open reading frame, ORF)。ORF1编码6个非结构蛋白,从N端到C端依次为N末端蛋白(p48)、核苷三磷酸酶(nucleoside triphosphatase, NTPase)、3A样蛋白(p22)、VPg(病毒基因组连接蛋白)、3C样蛋白酶(3CLpro)和RNA依赖的RNA聚合酶(RNA-dependent RNA polymerase, RdRp);ORF2和ORF3分别编码主要和次要衣壳蛋白:VP1和VP2[2]。HuNoV基因组的5′端共价连接了一个病毒蛋白VPg,研究表明它与病毒的翻译起始有关[3-4]。HuNoV是全球范围内引起人急性胃肠炎的主要病毒性病原[5],感染症状为呕吐、腹泻和发热等,传播方式为粪-口传播[6]。健康人感染后症状一般表现为自限性,但对于免疫缺陷者、老人和儿童,则可能导致严重后果,甚至危及生命[7-10]。
HuNoV于1972年被报道发现,至今已有四十多年[11],但其复制和致病机制仍不清楚,药物开发和疫苗研究也受到很大阻碍,主要原因之一在于缺乏合适高效的体外复制细胞模型[10, 12-14]。Mary K. Estes教授等应用EF-1α启动子表达系统构建了HuNoV GⅡ.3 U201感染性克隆,在多种细胞中检测到病毒复制,此构建策略也支持鼠诺如病毒(murine norovirus,MuNoVs)感染性克隆的复制,但是尝试用其来构建其他HuNoV感染性克隆时,RNA复制效率降低90%~99.9%[15],提示EF-1α启动子表达系统构建策略可能需要进一步优化,或者需要寻找更合适的HuNoV序列,这样才能实现高效复制。
鉴于此,本研究对EF-1α启动子表达系统构建的HuNoV GⅡ.3 U201感染性克隆是否能够复制进行验证,并与T7启动子表达系统构建策略相比较。为在临床病毒株中寻找更优序列,在EF-1α启动子表达系统中插入人工合成的HuNoV GⅡ.4临床病毒株全基因组片段,并在细胞中进行转染验证。期望本研究能为下一步优化HuNoV反向遗传体系进而构建高水平复制的体外培养系统提供新的思路。
1 材料与方法 1.1 材料 1.1.1 细胞和质粒肝癌细胞系Huh 7为本室保存[16],非洲绿猴肾成纤维细胞系COS 7(SV40转化)购自中国科学院上海生命科学院。HuNoV GⅡ.4临床粪便标本为上海市公共卫生临床中心保存。HuNoV GⅡ.3U201基因组序列来自GenBank,编号为AB039782[15],经体外合成后克隆于pLC-Zero-blunt, 病毒序列5′端添加T7启动子序列,得到的质粒命名为pU201。在病毒的RNA聚合酶活性位点GDD中引入突变,构建病毒RNA聚合酶失活突变质粒,得到的质粒命名为pU201-GNN。T7 RNA聚合酶编码序列经体外合成后克隆于phCMV载体,得到质粒phCMV-T7pol。依据文献报道[15],将U201序列克隆于带有EF-1α启动子的质粒,得到pEF-1α-U201与pEF-1α-U201-GNN。将Nluc基因同框融合于ORF1的N端,分别得到pU201-Nluc、pU201-Nluc-GNN、pEF-1α-U201-Nluc、pEF-1α-U201-Nluc-Δpol等质粒。在ORF2中替换一段序列,得到pEF-1α-U201-ORF2-Nluc、pEF-1α-U201-ORF2-Nluc-GNN。HuNoV GⅡ.4的序列经体外合成后克隆于带有EF-1α启动子的质粒,命名为PCTc,同时构建病毒RNA聚合酶活性位点失活的质粒PCTc-GNN及PCTc-GAA。所有质粒均经过Sanger测序进行验证。
1.1.2 试剂主要试剂有:DMEM培养基(美国Corning Cellgro公司,10-013-CVa),胎牛血清(以色列Biological Industries公司,04-001-1ACS),TRIzol Reagent(美国Invitrogen公司,15596018),TRIzol LS Reagent(美国Invitrogen公司,10296028),一步法荧光定量聚合酶链反应(polymerase chain reaction, PCR)试剂盒(日本TaKaRa公司,AK11417A),DNA转染试剂盒(美国Mirus公司,MIR 2300),RNA转染试剂盒(美国Mirus公司,MIR 2250),DNA消化酶试剂盒(日本TaKaRa公司,2270A),RNA酶抑制剂(日本TaKaRa公司,2313A),Opti-MEM Ⅰ减血清培养基(美国Invitrogen公司,31985-070),磷酸盐缓冲液(phosphate buffered saline,PBS;以色列Biological Industries公司,2030097),2′-C-甲基胞嘧啶核苷(2-CMC;美国MedChemExpress公司,HY-10468),RAN抽提试剂盒(德国QIAGEN公司,74104)。
1.2 方法 1.2.1 细胞培养人肝癌细胞系Huh 7、非洲绿猴肾成纤维细胞系COS 7均用含10%胎牛血清的DMEM培养基,于37 ℃、5% CO2恒温恒湿培养箱中培养,每2~3 d进行细胞传代。
1.2.2 质粒转染将Huh 7细胞或COS 7细胞按2.5×105 cells/mL均匀铺于48孔板中,每孔250 μL,16~18 h后进行转染。转染前每孔更换为220 μL新鲜培养液,按照每孔0.3 μg质粒转染。将0.3 μg质粒加到30 μL opti-MEM中,然后按照质粒与转染试剂LT1比例为1∶3(W/V)加入转染试剂,充分混匀后室温静置15~20 min,再将其均匀滴加到细胞中,晃匀后置于37 ℃、5% CO2恒温恒湿培养箱中培养。转染后8 h用PBS洗6遍,更换新鲜培养液,然后在不同时间点收集上清液或细胞样品。其他规模的孔板按照底面积调整转染质粒、opti-MEM和转染试剂的量。
1.2.3 总RNA抽提与RNA转染按QIAGEN RNeasy Mini Kit的步骤抽提后续转染用的总RNA,用NanoDrop 2000分光光度计定量后分装储存于-80 ℃。将Huh 7细胞或COS 7细胞按2.5×105 cells/mL的密度均匀铺于48孔板中,每孔250 μL,16~18 h后进行转染。转染前每孔更换为220 μL新鲜培养液,48孔板按照每孔0.3 μg RNA转染。将0.3 μg RNA加到30 μL opti-MEM中,再按RNA∶boost∶mRNA reagent为1∶2∶2(W/V/V)的比例依次加入转染试剂,充分混匀后室温静置2~5 min,随后将其均匀滴加到孔板中,晃匀后置于37 ℃、5% CO2恒温恒湿培养箱中培养。转染后6 h用PBS洗6遍,更换新鲜培养液,然后在不同时间点收集细胞样品。其他规模的孔板按照底面积调整转染质粒、opti-MEM和转染试剂的量。
1.2.4 实时荧光定量PCR收集细胞样品,用TRIzol裂解细胞后按TRIzol∶氯仿为1∶5(V/V)加入氯仿。收集上清液样品,用TRIzol LS裂解上清液,按上清液∶TRIzol LS∶氯仿为1∶3∶1(V/V/V)加入氯仿,充分混匀后12 800×g离心15 min,取上清液加入等体积的异丙醇(isopropyl alcohol),温和颠倒数次后室温静置15 min,12 800×g离心15 min。用预冷的75%乙醇洗涤沉淀物后于室温晾干,加入10 μL去RNA酶的焦炭酸二乙酯(diethyl pyrocarbonate,DEPC)水溶解。再将RNA用DNA酶消化处理,当RNA量在20~50 μg范围,按DNase Ⅰ∶RNase inhibitor为4∶1(V/V)依次加入DNA酶和RNA酶抑制剂,然后用水补足到50 μL,充分混匀后37 ℃水浴2 h,用DEPC水补足到100 μL,用TRIzol LS抽提RNA。最后取RNA,按照一步法荧光定量PCR试剂盒说明书进行RT-qPCR。定量PCR引物和探针见表 1。
| HuNoVs | GAPDH | |
| Forward primer | CARGARBCNATGTTYAGRTGGATGAG | GGTATCGTGGAAGGACTCATGAC |
| Reverse primer | TCGACGCCATCTTCATTCACA | ATGCCAGTGAGCTTCCCGTTCAGC |
| Probe | TPTGGGAGGGCGATCGCAATCT | CAATGCCACCCAGAAGACTGTGGATGGC |
收集细胞样品,在48孔板中每孔加入60 μL 1×passive lysis buffer,静置15 min以完全裂解后充分混匀,吸取20 μL混合液。然后开启荧光检测仪GloMaxⓇ 20/20 Luminometer (美国Promega公司),将100×底物用renilla luciferase buffer稀释,充分混匀。加入50 μL底物到待检测样品中,吹吸混匀后放置于仪器样品槽中读取荧光数值。
1.3 统计学分析应用Graphpad Prism 6软件来处理数据作图,并进行数据分析。采用非配对t检验进行组间差异比较,P < 0.05表明差异具有统计学意义。
2 结果 2.1 T7启动子和EF-1α启动子构建的HuNoV GⅡ. 3 U201的全长感染性克隆在细胞中短暂复制为验证T7启动子与EF-1α启动子构建的HuNoV全长克隆能否实现复制,人工合成含有T7启动子和EF-1α启动子的HuNoV GⅡ.3 U201病毒全长基因组片段,分别插入到载体中,将构建的质粒分别命名为pU201和pEF-1α-U201。在此基础上,通过将RdRp复制活性位点的氨基酸GDD突变成GNN,分别构建病毒RNA聚合酶失活的突变质粒。在COS 7细胞和Huh 7细胞中分别共转染T7聚合酶与T7启动子构建质粒以及单独转染EF-1α启动子构建质粒,于转染后不同时间点用TRIzol裂解细胞,用RT-qPCR检测细胞内HuNoV RNA水平。结果显示,pEF-1α-U201和pU201的病毒RNA水平随着时间增加逐渐升高,转染后第2天达到最大值,相较于复制活性位点发生突变的阴性对照,前者升高近3倍,后者升高约2倍。但第3天病毒复制水平显著下降,第4天与阴性对照水平一致(图 1B~1E)。结果提示,EF-1α启动子表达系统中病毒复制水平比T7启动子表达系统中的略高,但两种表达体系中病毒RNA皆不能长期复制。
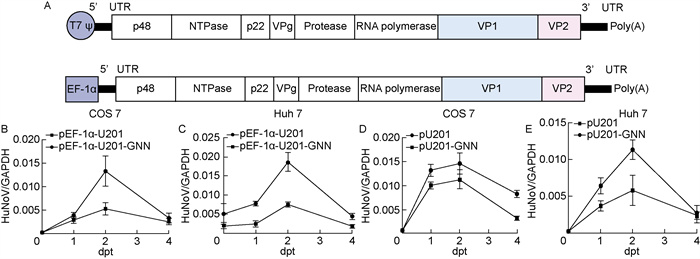
|
| A: Schematic diagrams of pU201 and pEF-1α-U201. pU201 is constructed with T7 promoter, and pU201-GNN has a mutation of active site in RNA polymerase. In the same way, pEF-1α-U201 is constructed with EF-1α promoter, and pEF-1α-U201-GNN has a mutation of active site in RNA polymerase. B-E: HuNoV RNA levels in transfected cells were determined by RT-qPCR and normalized against GAPDH RNA levels. Results are reported as mean±s (n=3). 图 1 HuNoV GⅡ. 3 U201的全长感染性克隆在细胞中短暂复制 Fig. 1 Infectious clones of HuNoV GⅡ. 3 U201 could replicate transiently in cells |
为便于检测HuNoV GⅡ. 3 U201复制水平,在pU201和pEF-1α-U201质粒的病毒基因组上游插入Nluc荧光素酶基因,分别命名为pU201-Nluc和pEF-1α-U201-Nluc(图 2A)。在COS 7细胞和Huh 7细胞中分别共转染T7聚合酶与T7启动子构建质粒以及单独转染EF-1α启动子构建质粒,转染后8 h用作用于病毒复制聚合酶的抑制剂2-CMC处理,然后于不同时间点收集细胞,检测Nluc荧光素酶活性。结果显示,pEF-1α-U201-Nluc和pU201-Nluc的Nluc荧光素酶活性随着时间增加而升高,转染后第2天达到最高值,与阴性对照以及抑制剂处理组相比,增高约2倍,相比于T7启动子构建的质粒增高更明显(图 2B~2E)。此外,pEF-1α-U201-Nluc在Huh 7细胞中的荧光素酶活性明显高于在COS 7细胞中,提示Huh 7细胞可能更有利于HuNoV复制。
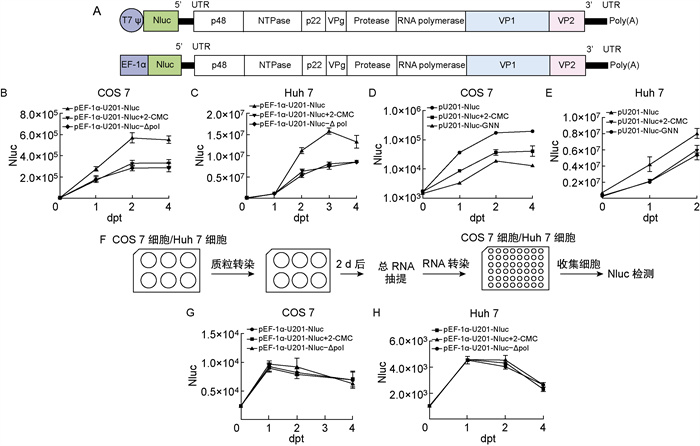
|
| A: Schematic diagrams of pU201-Nluc and pEF-1α-U201-Nluc. pU201-Nluc and pEF-1α-U201-Nluc are constructed basing on pU201 and pEF-1α-U201 respectively, and inserted NanoLucTM luciferase gene in front of the virus genome. pU201-Nluc-GNN has a mutation of active site in RNA polymerase based on pU201-Nluc, and pEF-1α-U201-Nluc-Δpol has a deletion of active site in RNA polymerase based on pEF-1α-U201-Nluc. B-E: NanoLucTM luciferase (Nluc) activity in the transfected cells. Huh 7 cells and COS 7 cells were transfected with pU201-Nluc, pU201-Nluc-GNN, pEF-1α-U201-Nluc, pEF-1α-U201-Nluc-Δpol, and another control group was added with 2-CMC. At different time post transfection, the cells were harvested. H: Scheme of experimental route to verify whether the infectious clones with Nluc reporter gene could achieve seconed-cycle replication. G-H: NanoLucTM luciferase (Nluc) activity in the transfected cells. Cells were transfected with total RNA extracted from cells transfected with pEF-1α-U201-Nluc and pEF-1α-U201-Nluc-Δpol. 图 2 带Nluc报告基因的HuNoV GⅡ. 3 U201全长感染性克隆在细胞中能实现一个周期复制 Fig. 2 Full-length infectious clones of HuNoV GⅡ. 3 U201 with Nluc reporter gene could achieve one cycle replication in cells |
为进一步验证含有Nluc报告基因的全长克隆形成的RNA能否复制,在COS 7细胞和Huh 7细胞中转染pEF-1α-U201-Nluc,2 d后抽提细胞总RNA,分别转染COS 7细胞和Huh 7细胞,在不同时间点收集细胞,检测Nluc荧光素酶活性(图 2F)。结果显示,与阴性对照以及抑制剂处理组相比,3组Nluc荧光素酶活性没有明显差异(图 2G~2H)。
2.3 EF-1α启动子构建的带Nluc报告基因的亚基因组克隆在Huh7细胞中实现一个周期复制为进一步验证EF-1α启动子表达系统构建的HuNoV GⅡ. 3 U201的亚基因组能否复制,将主要衣壳蛋白VP1部分序列替换成Nluc报告基因序列,命名为pEF-1α-U201-ORF2-Nluc(图 3A)。将质粒分别转染COS 7细胞和Huh 7细胞,于转染后不同时间点收集细胞,检测Nluc荧光素酶活性。结果显示,在COS 7细胞中各组荧光素酶活性无显著差异(图 3B)。与阴性对照以及抑制剂处理组相比,pEF-1α-U201-ORF2-Nluc的Nluc荧光素酶活性在Huh 7细胞中随着时间增加而升高,转染后第3天达到最高值,升高约2倍(图 3C)。
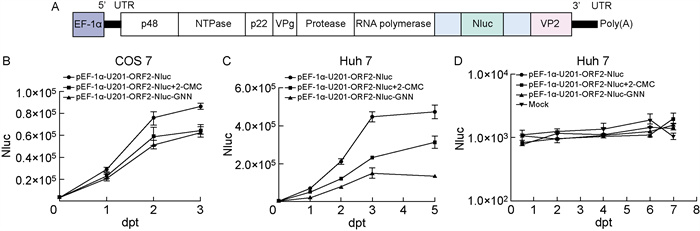
|
| A: Schematic diagrams of pEF-1α-U201-ORF2-Nluc. pEF-1α-U201-ORF2-Nluc is constructed basing on pEF-1α-U201, and a part of sequence of ORF2 is replaced by NanoLucTM luciferase (Nluc) gene. pEF-1α-U201-ORF2-Nluc-GNN has a mutation of active site in RNA polymerase. B-C: Nluc activity in the transfected cells. Cells were transfected with pEF-1α-U201-ORF2-Nluc, pEF-1α-U201-ORF2-Nluc-GNN, and another control group was added with 2-CMC. At different time post transfection, the cells were harvested. D: Nluc activity in the transfected cells. Cells were transfected with total RNA extracted from cells transfected with pEF-1α-U201-ORF2-Nluc, pEF-1α-U201-ORF2-Nluc-GNN, and another control group was added with 2-CMC. 图 3 EF-1α启动子构建的带Nluc报告基因的亚基因组克隆在Huh 7细胞中实现一个周期复制 Fig. 3 Subgenomic replicon with Nluc reporter gene constructed with EF-1α promoter could achieve a round of replication in Huh 7 cells |
为进一步探究pEF-1α-U201-ORF2-Nluc形成的病毒RNA在Huh 7细胞中能否复制,在Huh 7细胞中转染pEF-1α-U201-ORF2-Nluc,2 d后抽提细胞总RNA,并转染Huh 7细胞,于转染后不同时间点收集细胞,检测Nluc荧光素酶活性。结果显示,与阴性对照、抑制剂处理组以及空白对照组相比,不同时间点4组Nluc荧光素酶活性皆没有明显差异(图 3D)。
2.4 EF-1α启动子构建的HuNoV GⅡ.4临床病毒株全长感染性克隆不能复制为探究EF-1α启动子表达系统是否适用于不同HuNoV病毒株,挑选HuNoV GⅡ.4临床病毒株,人工合成HuNoV GⅡ.4临床病毒株全基因组片段,将其插入到含EF-1α启动子的载体中,构建质粒并命名为PCTc(图 4A)。同时在RdRp复制活性位点处进行核酸突变,构建复制缺陷质粒PCTc-GNN和PCTc-GAA作为阴性对照。在COS 7细胞和Huh 7细胞中分别转染上述质粒,不同时间点收集上清液,并于转染后2 d收集细胞,用RT-qPCR检测上清液中以及细胞内HuNoV RNA水平。结果显示,在上清液(图 4B~4C)以及细胞(图 4D~4E)中,与阴性对照以及抑制剂处理组相比,HuNoV RNA水平没有明显差异,表明EF-1α启动子构建的HuNoV GⅡ.4临床病毒株全长感染性克隆不能实现复制,提示HuNoV感染性克隆能否复制可能与病毒株相关。
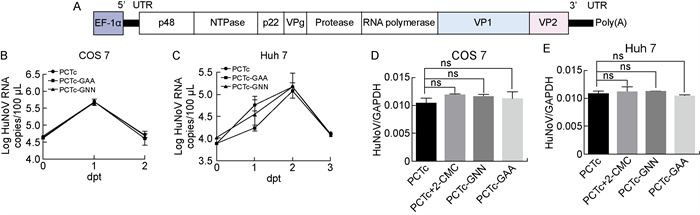
|
| A: Schematic diagrams of PCTc constructed with EF-1α promoter and HuNoV GⅡ.4 genome sequence from a Taiwan patient. PCTc-GNN and PCTc-GAA are constructed based on PCTc with a different mutation of active site in RNA polymerase, respectively. B-C: HuNoV RNA levels in 100 μL supernatants were determined by RT-qPCR. D-E: HuNoV RNA levels in transfected cells (Huh 7 cells and COS 7 cells) were determined by RT-qPCR and normalized against GAPDH RNA levels. Results are reported as mean±s(n=3). 图 4 EF-1α启动子表达体系构建的HuNoV GⅡ.4临床病毒株全长感染性克隆在细胞中不能复制 Fig. 4 4 Full-length infectious clone of HuNoV GⅡ.4 clinical strain constructed with EF-1α promoter could not replicate in cells |
HuNoV是全球范围内引起人急性胃肠炎的主要病毒性病原体,每年造成约7亿人感染和二十多万人死亡[8]。但是由于缺乏合适高效的体外培养细胞和动物模型,HuNoV的复制和致病机制还不甚清楚,也阻碍了药物和疫苗的研发。近年来有研究报道,利用EF-1α启动子表达系统构建的HuNoV GⅡ.3 U201反向遗传体系可在多种细胞中检测到病毒RNA复制和蛋白表达,还支持MuNoV感染性克隆的复制,但用其构建HuNoV其他基因型感染性克隆没有成功[15]。
本研究首先对已报道可复制的HuNoV GⅡ.3 U201的复制体系进行验证,同时与T7启动子表达系统[17]构建策略进行复制效率的比较。结果表明,EF-1α启动子表达系统所构建的全长感染性克隆以及亚基因组克隆均能实现2~3倍的复制,相较于T7启动子构建策略复制效率更高,说明不同启动子的构建策略可能会影响病毒的复制及复制水平。并且在不同宿主细胞,如Huh 7细胞和COS 7细胞中,HuNoV GⅡ.3 U201的全长克隆和亚基因组克隆的复制效率也有差异。其次,在质粒转染2 d后抽提转染细胞的总RNA再次进行RNA转染时,Nluc荧光素酶活性与对照组无明显差异,表明其并不能实现复制。这其中的原因一方面可能是细胞内天然免疫对病毒复制产生限制,导致病毒RNA复制效率低;另一方面抽提出的总RNA中HuNoV RNA含量太低,而且抽提出的HuNoV RNA可能不具有复制能力。目前,在世界范围内广泛流行的HuNoV基因型是GⅡ.4,但尚无能高效复制的HuNoV GⅡ.4体外复制系统。本研究挑选1株来自中国台湾患者的HuNoV GⅡ.4临床病毒株,人工合成其全基因组片段,采用EF-1α启动子表达系统构建其全长基因组克隆,并在Huh 7和COS 7细胞中进行验证。结果显示,与对照组相比,HuNoV RNA水平无明显差异,表明其并不能实现复制。结果提示,在构建HuNoV反向遗传体系时,不同病毒序列的复制能力有差异。
宿主因素也可能影响HuNoV的复制效率。已有研究报道,在干细胞来源的非转化人肠道类肠状细胞(human intestinal enteroid, HIE)中成功培养多种HuNoV毒株,但是有些基因型的HuNoV复制具有胆汁依赖性,需要额外添加胆汁才能实现感染复制或者提高复制效率[18-19]。此外,也有研究报道,多种基因型的HuNoV能够在斑马幼鱼中实现高效复制,且复制水平能维持至少6 d[20]。以上研究提示可以尝试在类器官以及斑马幼鱼中进行HuNoV反向遗传体系的验证,同时也可以进一步探究宿主对其复制效率的影响。
综上所述,本研究结果提示EF-1α启动子表达系统可能相比T7启动子更优,但所构建的克隆在细胞中的复制效率皆低,且不能实现持续复制,其复制能力也可能与不同序列的病毒株相关。关于HuNoV的培养模型还在不断研究中,目前仍没有高效可长期复制的细胞和动物培养模型。下一步将继续优化HuNoV反向遗传体系构建策略,构建不同序列的HuNoV临床病毒株感染性克隆,进而在细胞、类器官以及斑马幼鱼中进行验证,期望建立能够高水平复制的HuNoV体外培养体系,进而为研究HuNoV复制机制提供有力的工具。
| [1] |
Duizer E, Schwab KJ, Neill FH, Atmar RL, Koopmans MPG, Estes MK. Laboratory efforts to cultivate noroviruses[J]. J Gen Virol, 2004, 85(Pt 1): 79-87.
[URI]
|
| [2] |
Vinjé J. Advances in laboratory methods for detection and typing of norovirus[J]. J Clin Microbiol, 2015, 53(2): 373-381.
[DOI]
|
| [3] |
Guix S, Asanaka M, Katayama K, Crawford SE, Neill FH, Atmar RL, Estes MK. Norwalk virus RNA is infectious in mammalian cells[J]. J Virol, 2007, 81(22): 12238-12248.
[DOI]
|
| [4] |
Chang KO, Sosnovtsev SV, Belliot G, King AD, Green KY. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line[J]. Virology, 2006, 353(2): 463-473.
[DOI]
|
| [5] |
Ramani S, Atmar RL, Estes MK. Epidemiology of human noroviruses and updates on vaccine development[J]. Curr Opin Gastroenterol, 2014, 30(1): 25-33.
[DOI]
|
| [6] |
Moore MD, Goulter RM, Jaykus LA. Human norovirus as a foodborne pathogen: challenges and developments[J]. Annu Rev Food Sci Technol, 2015, 6: 411-433.
[DOI]
|
| [7] |
Green KY. Norovirus infection in immunocompromised hosts[J]. Clin Microbiol Infec, 2014, 20(8): 717-723.
[DOI]
|
| [8] |
Trivedi TK, Desai R, Hall AJ, Patel M, Parashar UD, Lopman BA. Clinical characteristics of norovirus-associated deaths: a systematic literature review[J]. Am J Infect Control, 2013, 41(7): 654-657.
[DOI]
|
| [9] |
Yu JM, Liang ZY, Guo K, Sun XM, Zhang Q, Dong YJ, Duan ZJ. Intra-host evolution of norovirus GII4 in a chronic infected patient with hematopoietic stem cell transplantation [J]. Front Microbiol, 2020, 11: 375.
[DOI]
|
| [10] |
de Graaf M, van Beek J, Koopmans MP. Human norovirus transmission and evolution in a changing world[J]. Nat Rev Microbiol, 2016, 14(7): 421-433.
[DOI]
|
| [11] |
Kapikian AZ, Wyatt RG, Dolin R, Thornhill TS, Kalica AR, Chanock RM. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis[J]. J Virol, 1972, 10(5): 1075-1081.
[DOI]
|
| [12] |
Lay MK, Atmar RL, Guix S, Bharadwaj U, He H, Neill FH, Sastry KJ, Yao Q, Estes MK. Norwalk virus does not replicate in human macrophages or dendritic cells derived from the peripheral blood of susceptible humans[J]. Virology, 2010, 406(1): 1-11.
[DOI]
|
| [13] |
Straub TM, H öner zu Bentrup K, Orosz-Coghlan P, Dohnalkova A, Mayer BK, Bartholomew RA, Valdez CO, Bruckner-Lea CJ, Gerba CP, Abbaszadegan M, Nickerson CA. In vitro cell culture infectivity assay for human noroviruses[J]. Emerg Infect Dis, 2007, 13(3): 396-403.
[DOI]
|
| [14] |
Jones MK, Grau KR, Costantini V, Kolawole AO, de Graaf M, Freiden P, Graves CL, Koopmans M, Wallet SM, Tibbetts SA, Schultz-Cherry S, Wobus CE, Vinjé J, Karst SM. Human norovirus culture in B cells[J]. Nat Protoc, 2015, 10(12): 1939-1947.
[DOI]
|
| [15] |
Katayama K, Murakami K, Sharp TM, Guix S, Oka T, Takai-Todaka R, Nakanishi A, Crawford SE, Atmar RL, Estes MK. Plasmid-based human norovirus reverse genetics system produces reporter-tagged progeny virus containing infectious genomic RNA[J]. Proc Natl Acad Sci U S A, 2014, 111(38): E4043-E4052.
[DOI]
|
| [16] |
Yi Z, Pan T, Wu X, Song W, Wang S, Xu Y, Rice CM, Macdonald MR, Yuan Z. Hepatitis C virus co-opts Ras-GTPase-activating protein-binding protein 1 for its genome replication[J]. J Virol, 2011, 85(14): 6996-7004.
[DOI]
|
| [17] |
Katayama K, Hansman GS, Oka T, Ogawa S, Takeda N. Investigation of norovirus replication in a human cell line[J]. Arch Virol, 2006, 151(7): 1291-1308.
[DOI]
|
| [18] |
Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinjé J, Tibbetts SA, Wallet SM, Karst SM. Enteric bacteria promote human and mouse norovirus infection of B cells[J]. Science, 2014, 346(6210): 755-759.
[DOI]
|
| [19] |
Estes MK, Ettayebi K, Tenge VR, Murakami K, Karandikar U, Lin SC, Ayyar BV, Cortes-Penfield NW, Haga K, Neill FH, Opekun AR, Broughman JR, Zeng XL, Blutt SE, Crawford SE, Ramani S, Graham DY, Atmar RL. Human norovirus cultivation in nontransformed stem cell-derived human intestinal enteroid cultures: success and challenges[J]. Viruses, 2019, 11(7): 638-650.
[DOI]
|
| [20] |
Van Dycke J, Ny A, Concei ç ão-Neto N, Maes J, Hosmillo M, Cuvry A, Goodfellow I, Nogueira TC, Verbeken E, Matthijnssens J, de Witte P, Neyts J, Rocha-Pereira J. A robust human norovirus replication model in zebrafish larvae[J]. PLoS Pathog, 2019, 15(9): e1008009.
[DOI]
|
 2021, Vol. 16
2021, Vol. 16


