具核梭杆菌(Fusobacterium nucleatum)是人类口腔及肠道黏膜的共生菌,因其可导致包括牙周炎、牙髓炎在内的多种口腔炎症疾病,最初作为口腔机会致病菌被研究。近年来,随着肠道微生态学研究的升温,具核梭杆菌与结直肠癌(colorectal cancer, CRC)的关系研究受到了更多关注,大量研究发现具核梭杆菌在CRC患者的肠组织或粪便样品中检出丰度显著增加,表明具核梭杆菌与CRC的发生发展具有关联性,且有相关基础研究证明其对于CRC能够起到“驱动”作用。健康状态下,人体肠道微生态系统呈稳态,微生物群与人体的肠道黏膜免疫系统维持动态平衡;疾病状态下,正常肠道微生物群结构被破坏,而具核梭杆菌丰度的增加会进一步促进CRC的发生发展[1]。目前发现具核梭杆菌影响CRC进程的机制包括调节肿瘤微环境、刺激肠道慢性炎症等。本文对CRC导致具核梭杆菌富集的证据进行总结,并对具核梭杆菌促进CRC发生发展的机制进行综述。
1 具核梭杆菌的丰度在CRC患者中显著增加Koliarakis等[1]通过16S rRNA测序和宏基因组测序检测到CRC肿瘤组织中的具核梭杆菌丰度相对正常结直肠组织更高。在另一项研究中,Sunny等[2]也通过定量聚合酶链式反应(quantitative polymerase chain reaction, qPCR)从CRC患者粪便中检出具核梭杆菌,阳性率为72.12%(75/104)。Li等[3]从CRC患者冷冻组织和福尔马林固定石蜡包埋(Formalin-Fixed and Partffin-Embedded, FFPE)组织中检出具核梭杆菌的阳性率为87.13%(88/101)。Tahara等[4]用qPCR对149例原发性CRC组织标本中的具核梭杆菌进行鉴定,发现有74.50%(111/149)的CRC组织样本呈具核梭杆菌阳性。Park等[5]在160例经手术切除的微卫星高度不稳定性(microsatellite instability-high, MSI-H)分子亚型CRC的组织中,通过qPCR测定具核梭杆菌16S rRNA基因序列,发现MSI-H表型的CRC组织中,具核酸杆菌丰度增加与巨噬细胞浸润增多、CDKN2A基因启动子甲基化均显著相关。杨垒等[6]收集了35例大肠癌及其癌旁组织标本, 应用qPCR检测具核梭杆菌在肠癌及癌旁组织的丰度, 发现具核梭杆菌在肠癌组织中呈高感染负荷状态, 且主要定殖于大肠癌黏膜表面。表 1展示了人体研究中具核梭杆菌与CRC相关性的研究证据。
| Author | Positive cases, n(%) | Test method | Test sample |
| Tahara et al.[4] | 111 (74.50) | qPCR | Tissue samples |
| Mima et al.[8] | 134 (12.54) | qPCR | Tissue samples |
| Yang et al.[6] | 27 (77.14) | qPCR | Tissue samples |
| Park et al.[5] | 107 (66.88) | qPCR | Tissue samples |
| Li et al.[3] | 88 (87.13) | FISH & FQ-PCR | Frozen tissue &FFPE |
| Ito et al.[9] | 286 (56.97) | qPCR | FFPE |
| Sukawa et al..[10] | 76 (12.71) | qPCR | FFPE |
| Nosho et al.[11] | 44 (8.61) | qPCR | FFPE |
| Sunny et al.[2] | 75 (72.12) | qPCR | Faeces |
| Suehiro et al.[12] | 85 (53.80) | ddPCR | Faeces |
| qPCR:Quantitative polymerase chain reaction;FQ-PCR:Fluorescence quantitative PCR;ddPCR:Drop like digital PCR;FISH:fluorescence in situ hybridization;FFPE:Formalin fixed paraffin embedding | |||
为了确定具核梭杆菌感染是否是结直肠癌发生的早期事件,在Flanagan等[7]对52例爱尔兰患者的结肠腺瘤组织中的具核梭杆菌进行了检测,发现这些结直肠腺瘤患者的具核梭杆菌水平在病变组织和正常组织中无显著性差异(P=0.06),而在高度不典型增生组织中的水平显著高于正常组织(P=0.015)。所以当结直肠腺瘤组织从轻、中度不典型增生发展到高度不典型增生时,具核梭杆菌的数量出现了明显的增加。
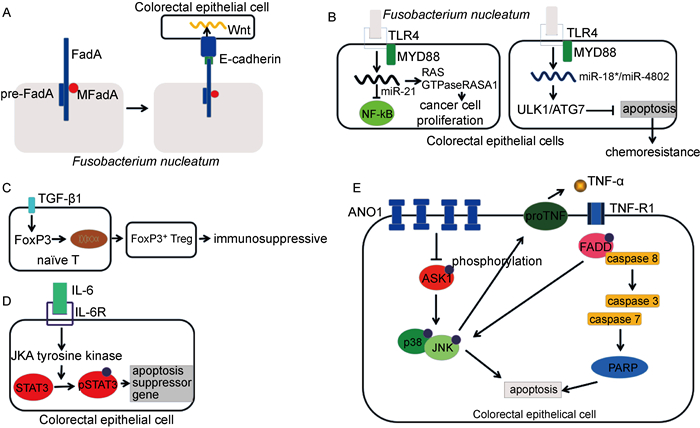
2 具核梭杆菌促进CRC发生的分子机制 2.1 通过FadA/E-钙粘蛋白作用黏附并侵入结、直肠上皮细胞在具核梭杆菌黏附并侵入结肠上皮细胞的过程中,最主要的是FadA与E-钙粘蛋白的相互作用。FadA是具核梭杆菌中高度保守的蛋白[13]。它存在两种形式,一种是不分泌的完整的前FadA(pre-FadA),另一种是由111个氨基酸组成的分泌型成熟FadA(MFadA)[1]。pre-FadA与MFadA结合形成活性复合物FadAc [14]。具核梭杆菌与细胞膜结合后,与E-钙粘蛋白上EC5结构域中的一段区域结合,并与E-钙粘蛋白内化(如图 1A所示)。此外,FadA与E-钙粘蛋白的相互作用足以激活Wnt和癌基因[15]。
|
| A: FadA forms the structure of microfilament similar to the pili, and MFadA binds to the base of microfilament to extend the microfilament through the outer membrane. Then pre-FadA binds to the base to terminate the extension of microfilament. FadA binds to E-cadherin on the surface of colorectal epithelial cells to complete adhesion. FadA modulates E-cadherin and activates β-catenin signaling, leading to increased expression of Wnt genes. B: By activating TLR4-MyD88 pathway, Fusobacterium nucleatum can up-regulate miR21, activate NF-κB, inhibit the expression of Ras GTPase-actvating protein 1(RASA1), and thus promote the proliferation of colorectal cancer cells. By activating TLR4-MyD88 signal, Fusobacterium nucleatum can inhibit the expression of miR-18a* and miR-4802, induce the expression of phosphorylated ULK1, ULK1 and autophagy protein, and thus activate the resistance to chemotherapy. C: TGF-β1 up-regulated, induced Foxp3 transcription factor expression, promoted the differentiation of initial T cells to regulatory T cells, resulting in local immunosuppression. D: The binding of IL-6 and IL-6R results in the activation of JAK tyrosine kinase in the cytoplasm, and then the activation of STAT3. With the activation of STAT3, many genes, including bcl-xl, mcl-1, x2ap, c-myc and fas, have been changed to regulate the growth, differentiation and survival of cells. E: The overexpression of ANO1 can inhibit the phosphorylation of ASK1 (apoptosis signal regulating kinase 1), so as to inhibit the phosphorylation of P38 and JNK (c-Jun amino terminal kinase) and down-regulate TNF-α, finally inhibit the fas associated protein with a level death domain (FADD) and caspase family, and inhibit apoptosis. 图 1 具核梭杆菌促进CRC发生的分子机制图 Fig. 1 Molecular mechanism of promoting CRC by Fusobacterium nucleatum |
具核梭杆菌黏附并侵入细胞后,通过活化TLR4-MYD88信号,上调miR21表达,从而抑制核因子κB(nuclear factor-κB,NF-κB),并且提高了RAS GTP酶激活蛋白1(Ras GTPase-activating protein 1, RASA1)的水平,进而促进大肠癌细胞增殖[16]。同时,具核梭杆菌通过活化TLR4-MYD88信号抑制miR-18a*和miR-4802表达,诱导磷酸化uncoordinated-51-like kinase 1(ULK1)和自噬蛋白的表达,使得细胞具有化疗抗性[8](如图 1B所示)。
2.3 调节肿瘤微环境在与具核梭杆菌有关的CRC发生过程中,转化生长因子β1(transforming growth factor-β1, TGF-β1)是肿瘤免疫微环境中重要的炎症因子,其在肿瘤免疫微环境中与肿瘤免疫逃逸相关[17]。具核梭杆菌感染肠道后,促进TGF-β1表达上调,进而诱导FoxP3转录因子表达[18],促使初始T细胞向调节性T细胞(regulatory T cell, Treg)分化[19](见图 1C)。这与幽门螺杆菌(Helicobacter pylori)诱导产生Treg细胞导致免疫抑制进而导致胃癌的机制相似[20]。而且,具核梭杆菌中的Fap2蛋白与免疫受体酪氨酸抑制模体(immunoreceptor tyrosine-based inhibitory motif, ITIM)结合可抑制自然杀伤细胞对肿瘤细胞的杀伤活性[21],影响肿瘤免疫微环境,并且产生局部免疫抑制效应,促进肿瘤细胞逃避机体的免疫监视。
2.4 刺激肠道慢性炎症慢性炎症在激活肿瘤通路中发挥了重要作用,而白细胞介素(interleukin, IL-22)和IL-6最为关键[22-23]。有研究使用C57BL/6小鼠建立结、直肠癌和具核梭杆菌作用模型,结果显示,与PBS对照组相比,具核梭杆菌处理的小鼠黏膜组织中, 炎症因子IL-6,IL-22和STAT3通路激活标志p-STAT3的蛋白表达量显著增高。此结果提示,具核梭杆菌可能刺激小鼠肠道组织IL-6和IL-22的表达,继而激活p-STAT3通路(见图 1D),进而促进结、直肠癌的发生。该研究还证实,在IL-22和IL-6作用下,结、直肠癌细胞增殖能力显著提高,其中IL-22作用更明显[24]。IL-22主要由Th17和Th22等细胞分泌, 而这些免疫细胞迁移的重要信号是趋化因子CCL20(C-C motif ligand 20)对趋化因子受体CCR6的趋化作用[25]。前期研究发现, CCR6阳性T细胞在结、直肠癌患者黏膜环境中聚集, 在小鼠模型中验证了CCL20在黏膜上皮表达增高[26]。因此推测,具核梭杆菌一方面促进IL-22和IL-6的分泌,激活p-STAT3通路,促进结、直肠癌的发生,另一方面具核梭杆菌通过提高相关趋化因子受体的表达,促进上述作用。
2.5 提高CRC的抗化疗性人钙激活氯通道蛋白1(Anotamin-1,ANO1)定位于人染色体11q13区域[27],研究发现在许多癌症中,ANO1经常被高表达[28]。有研究发现具核梭杆菌侵入结肠上皮细胞后可诱导ANO1表达,进而抑制奥沙利铂和5-氟尿嘧啶诱导的细胞凋亡[29](见图 1E)。同时,具核梭杆菌激活TLR4和MYD88后,选择性下调miRNA-18a*和miRNA-4802的表达,继而导致ULK1和自噬相关蛋白7(autophagy-related protein 7, ATG7)表达上调激活自噬,进而引起CRC患者对化疗药物产生抗性[30](见图 1B)。
3 CRC导致具核梭杆菌感染的可能机制目前相关研究大多关注具核梭杆菌对于CRC发生发展的影响,并且证明具核梭杆菌对CRC具有明确的促进作用。但同时不能否认CRC的发病能够影响具核梭杆菌的感染或富集。自噬是一种高度保守的分解代谢过程,通过形成名为自噬体的双膜囊泡并螯合胞质成分,将其传递至溶酶体进行降解。肿瘤细胞的自噬作用在影响微环境中的微生物组成方面起着关键作用,而且有证据表明肿瘤细胞激活的自噬活性可能会消除细胞内的微生物[31-36]。Koichiro等[37]以724例大肠癌患者为研究对象发现,肿瘤BECN 1 (beclin 1)表达的自噬活性与大肠癌组织中具核梭杆菌的数量成反比关系,提示自噬可能会消除具核梭杆菌。而相应自噬活性变弱会导致具核梭杆菌在肠道组织的富集。
4 展望越来越多的证据显示,具核梭杆菌存在于肠道,其通过FadA /E-钙粘蛋白作用、激活TLR4、调节肿瘤微环境、刺激肠道慢性炎症等多方面作用于肠道,协同其他因素对肠道产生影响,诱导结、直肠癌的产生。这些作用机制可能提示新的防治结、直肠癌的研究方向和靶点。目前,结、直肠癌影响具核梭杆菌的机制尚未完全明确,揭示其对具核梭杆菌的影响可为探索结、直肠癌的治疗开辟新的研究领域,提供新的思路。今后,能够特异性针对具核梭杆菌的药物、噬菌体以及化合物仍值得进一步探索。
| [1] |
Koliarakis I, Messaritakis I, Nikolouzakis TK, Hamilos G, Souglakos J, Tsiaoussis J. Oral bacteria and intestinal dysbiosis in colorectal cancer[J]. Int J of Mol Sci, 2019, 20(17): 4146.
[DOI]
|
| [2] |
Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G, Lam TYT, Zhang L, Wu JCY, Chan FKL, Ng SSM, Wong MCS, Ng SC, Wu WKK, Yu J, Sung JJY. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia[J]. Gut, 2017, 66(8): 1441-1448.
[DOI]
|
| [3] |
Li YY, Ge QX, Cao J, Zhou YJ, Du YL, Shen B, Wan YJ, Nie YQ. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients[J]. World J Gastroenterol, 2016, 22(11): 3227-3233.
[DOI]
|
| [4] |
Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, An B, Shureiqi I, Toyota M, Kondo Y, Estécio MR, Issa JP. Fusobacterium in colonic flora and molecular features of colorectal carcinoma[J]. Cancer Res, 2014, 74(5): 1311-1318.
[DOI]
|
| [5] |
Park HE, Kim JH, Cho NY, Lee HS, Kang GH. Intratumoral Fusobacterium nucleatum abundance correlates with macrophage infiltration and CDKN2A methylation in microsatellite-unstable colorectal carcinoma[J]. Virchows Arch, 2017, 471(3): 329-336.
[DOI]
|
| [6] |
杨垒, 杨永志, 秦环龙. 大肠癌和癌旁组织中具核梭杆菌丰度差异的研究[J]. 同济大学学报(医学版), 2015, 36(5): 34-38. [CNKI]
|
| [7] |
Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, Jenab M, Prehn JHM, Hughes DJ. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome[J]. Eur J Clin Microbiol Infect Dis, 2014, 33: 1381-1390.
[DOI]
|
| [8] |
Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, Kostic AD, Giannakis M, Bullman S, Milner DA, Baba H, Giovannucci EL, Garraway LA, Freeman GJ, Dranoff G, Garrett WS, Huttenhower C, Meyerson M, Meyerhardt JA, Chan AT, Fuchs CS, Ogino S. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis[J]. Gut, 2016, 65(12): 1973-1980.
[DOI]
|
| [9] |
Ito M, Kanno S, Nosho K, Sukawa Y, Mistuhashi K, Kurihara H, Igarashi H, Takahashi T, Tachibana M, Takahashi M, Takahashi H, Yoshii S, Takenouchi T, Hasegawa T, Okita K, Hirata K, Maruyama R, Suzuki H, Imai K, Yamamoto H, Shinomura Y. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway[J]. Int J Cancer, 2015, 137(6): 1258-1268.
[DOI]
|
| [10] |
Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K, Kostic AD, Giannakis M, Watanabe H, Bullman S, Milner DA, Harris CC, Giovannucci E, Garraway LA, Freeman GJ, Dranoff G, Chan AT, Garrett WS, Huttenhower C, Fuchs CS, Ogino S. Fusobacterium nucleatum and T cells in colorectal carcinoma[J]. JAMA Oncol, 2015, 1(5): 653-661.
[DOI]
|
| [11] |
Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, Kanno S, Yamamoto I, Ishigami K, Igarashi H, Maruyama R, Imai K, Yamamoto H, Shinomura Y. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer[J]. World J Gastroenterol, 2016, 22(2): 557-566.
[DOI]
|
| [12] |
Suehiro Y, Sakai K, Nishioka M, Hshimoto S, Takami T, Higaki S, Shindo Y, Hazama S, Oka M, Nagano H, Sakaida I, Yamasaki T. Highly sensitive stool DNA testing of Fusobacterium nucleatum as a marker for detection of colorectal tumours in a Japanese population[J]. Ann Clin Biochem, 2017, 54(1): 86-91.
[DOI]
|
| [13] |
Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, Li M, Sojar HT, Genco RJ, Kuramitsu HK, Deng CX. Identification and characterization of a novel adhesin unique to oral fusobacteria[J]. J Bacteriol[J], 2005, 187(15): 5330-5340.
[DOI]
|
| [14] |
Xu M, Yamada M, Li M, Liu H, Chen SG, Han YW. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells[J]. J Biol Chem, 2007, 282(34): 25000-25009.
[DOI]
|
| [15] |
Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin[J]. Cell Host Microbe, 2013, 14(2): 195-206.
[DOI]
|
| [16] |
Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, Li H, Guo B, Zhu Q, Wei Q, Moyer MP, Wang P, Cai S, Goel A, Qin H, Ma Y. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of microRNA-21[J]. Gastroenterology, 2017, 152(4): 851-866.
[DOI]
|
| [17] |
Massagué J. TGFbeta in cancer[J]. Cell, 2008, 134(2): 215-230.
[DOI]
|
| [18] |
Curotto de Lafaille MA, Lafaille JJ. Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor[J]. Immunity, 2009, 30(5): 626-635.
[DOI]
|
| [19] |
Raitala A, Karjalainen J, Oja SS, Kosunen TU, Hurme M. Helicobacter pylori-induced indoleamine 2, 3-dioxygenase activity in vivo is regulated by TGFB1 and CTLA4 polymorphisms[J]. Mol Immunol, 2007, 44(5): 1011-1014.
[DOI]
|
| [20] |
Bagheri N, Azadegan-Dehkordi F, Rahimian G, Rafieian-Kopaei M, Shirzad H. Role of regulatory T-cells in different clinical expressions of Helicobacter pylori infection[J]. Arch Med Res, 2016, 47(4): 245-254.
[DOI]
|
| [21] |
Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklić K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack[J]. Immunity, 2015, 42(2): 344-355.
[DOI]
|
| [22] |
Yu YN, Yu TC, Zhao HJ, Sun TT, Chen HM, Chen HY, An HF, Weng YR, Yu J, Li M, Qin WX, Ma X, Shen N, Hong J, Fang JY. Berberine may rescue Fusobacterium nucleatum-induced colorectal tumorigenesis by modulating the tumor microenvironment[J]. Oncotarget, 2015, 6(31): 32013-32026.
[DOI]
|
| [23] |
Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3[J]. Nat Rev Cancer, 2009, 9(11): 798-809.
[DOI]
|
| [24] |
Lu SY, Yan TT, Guo FF, Chen YX, Hong J, Fang JY. Interleukin-22 and Fusobacterium nucleatum cooperate in promoting colorectal cancer (in Chinese)[J]. Sci Sin Vitae, 2018, 48(6): 684-691.
[DOI]
|
| [25] |
Zhuang Y, Peng LS, Zhao YL, Shi Y, Mao XH, Guo G, Chen W, Liu XF, Zhang JY, Liu T, Luo P, Yu PW, Zou QM. Increased intratumoral IL-22-producing CD4+T cells and Th22 cells correlate with gastric cancer progression and predict poor patient survival[J]. Cancer Immunol Immunother, 2012, 61(11): 1965-1975.
[DOI]
|
| [26] |
Kryczek I, Lin Y, Nagarsheth N, Peng D, Zhao L, Zhao E, Vatan L, Szeliga W, Dou Y, Owens S, Zgodzinski W, Majewski M, Wallner G, Fang J, Huang E, Zou W. IL-22+CD4+ T Cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L[J]. Immunity, 2014, 40(5): 772-784.
[DOI]
|
| [27] |
Katoh M, Katoh M. FLJ10261 gene, located within the CCND1-EMS1 locus on human chromosome 11q13, encodes the eight-transmembrane protein homologous to C12orf3, C11orf25 and FLJ34272 gene products[J]. Int J Oncol, 2003, 22(6): 1375-1381.
[URI]
|
| [28] |
Perez-Ordoñez B, Beauchemin M, Jordan RC. Molecular biology of squamous cell carcinoma of the head and neck[J]. J Clin Pathol, 2006, 59(5): 445-453.
[DOI]
|
| [29] |
Song Y, Gao J, Guan L, Chen X, Gao J, Wang K. Inhibition of ANO1/TMEM16A induces apoptosis in human prostate carcinoma cells by activating TNF-α signaling[J]. Cell Death Dis, 2018, 9(6): 703.
[DOI]
|
| [30] |
Yu T, Guo F, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang JY. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy[J]. Cell, 2017, 170(3): 548-563.
[DOI]
|
| [31] |
Zhang L, Hu W, Cho CH, Chan FK, Yu J, Fitzgerald JR, Cheung CK, Xiao ZG, Shen J, Li LF, Li MX, Wu JC, Ling TK, Chan JY, Ko H, Tse G, Ng SC, Yu S, Wang MH, Gin T, Ashktorab H, Smoot DT, Wong SH, Chan MT, Wu WK. Reduced lysosomal clearance of autophagosomes promotes survival and colonization of Helicobacter pylori[J]. J Pathol, 2018, 244(4): 432-444.
[DOI]
|
| [32] |
Heath RJ, Goel G, Baxt LA, Rush JS, Mohanan V, Paulus GLC, Jani V, Lassen KG, Xavier RJ. RNF166 determines recruitment of adaptor proteins during antibacterial autophagy[J]. Cell Rep, 2016, 17(9): 2183-2194.
[DOI]
|
| [33] |
Hubber A, Kubori T, Coban C, Matsuzawa T, Ogawa M, Kawabata T, Yoshimori T, Nagai H. Bacterial secretion system skews the fate of Legionella-containing vacuoles towards LC3-associated phagocytosis[J]. Sci Rep, 2017, 7: 44795.
[DOI]
|
| [34] |
Mimouna S, Bazin M, Mograbi B, Darfeuille-Michaud A, Brest P, Hofman P, Vouret-Craviari V. HIF1A regulates xenophagic degradation of adherent and invasive Escherichia coli (AIEC)[J]. Autophagy, 2014, 10(12): 2333-2345.
[DOI]
|
| [35] |
Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity[J]. Nat Rev Immunol, 2013, 13(10): 722-737.
[DOI]
|
| [36] |
Gomes LC, Dikic I. Autophagy in antimicrobial immunity[J]. Mol Cell, 2014, 54(2): 224-233.
[DOI]
|
| [37] |
Haruki K, Kosumi K, Hamada T, Twombly TS, Väyrynen JP, Kim SA, Masugi Y, Qian ZR, Mima K, Baba Y, da Silva A, Borowsky J, Arima K, Fujiyoshi K, Lau MC, Li P, Guo C, Chen Y, Song M, Nowak JA, Nishihara R, Yanaga K, Zhang X, Wu K, Bullman S, Garrett WS, Huttenhower C, Meyerhardt JA, Giannakis M, Chan AT, Fuchs CS, Ogino S. Association of autophagy status with amount of Fusobacterium nucleatum in colorectal cancer[J]. J pathol, 2020, 250(4): 397-408.
[DOI]
|
 2021, Vol. 16
2021, Vol. 16


