2. 复旦大学附属闵行医院重症医学科,上海 201199;
3. 复旦大学附属闵行医院急性损伤和继发感染研究与转化实验室,上海 201199
2. Intensive Care Unit, Minhang Hospital, Fudan University, Shanghai 201199, China;
3. Research and Translational Laboratory of Acute Injury and Secondary Infection, Minhang Hospital, Fudan University, Shanghai 201199, China
血流感染(bloodstream infection,BSI)对患者的生命是一种严重的威胁,是全球范围内的一个重大公共卫生问题[1]。BSI预后与诊断的实效性、准确性、抗菌药物运用的及时性密切相关[2]。据估计,全球血流感染的死亡人数达800万/年,每4 s就有1人死亡;每延迟治疗1 h,病死率就增加7.6%,延迟治疗6 h,病死率将增加58%[3]。
血流感染的主要病原体有大肠埃希菌、肺炎克雷伯菌、鲍曼不动杆菌、铜绿假单胞菌、金黄色葡萄球菌、肠球菌和念珠菌等[4]。早期明确感染的病原体可以指导临床医生及时有效地进行抗感染治疗,尤其是脓毒症休克的病人,80%的患者得到了挽救[5]。血培养(blood culture,BC)依然是目前诊断BSI的金标准,但其耗时长、采血量多、阳性率低、易污染,无法全面满足临床的需求[6]。
液滴数字PCR(droplet digital polymerase chain reaction PCR,ddPCR)是第3代PCR扩增技术,其原理是将一个荧光定量PCR反应体系分配到数万个反应单元中,每个反应单元中包含或不包含1个或多个拷贝的核酸分子,进行“单分子模板PCR扩增”,扩增结束后,通过阳性反应单元的数量和泊松分布的原理计算原始样本中核酸的拷贝数[7]。ddPCR具有灵敏度高、特异性强和绝对定量等优点,在快速鉴定病原体和抗生素耐药性(antimicrobial resistance,AMR)分析方面显示出巨大的潜力[8]。本研究采用液滴数字PCR对疑似血流感染的危重患者进行检测,并与传统的血培养结果进行多种参数的比较,以评估液滴数字PCR在诊断血流感染中的应用价值。
1 资料与方法 1.1 研究对象本研究为单中心前瞻性研究,经复旦大学附属闵行医院伦理委员会批准,伦理批件号:KY2021-790。筛选2021年12月—2023年8月入住复旦大学附属闵行医院重症监护病房的疑似血流感染的患者。
1.2 纳入和排除标准纳入标准:年龄≥18岁,符合Sepsis-1诊断标准的住院病人,即感染且至少具备以下2项指标:①体温>38 ℃或 < 36 ℃;②心率>90次/min;③呼吸频率>20次/min或动脉二氧化碳分压(PaCO2) < 32 mmHg(4.3 kPa)或机械通气;④外周血白细胞计数>12.0×109/L或 < 4.0×109/L,或未成熟粒细胞>10%。排除标准:临床信息不详者。收集患者的临床信息:性别、年龄、临床诊断、实验室结果。实验室结果包括血培养、降钙素原(procalcitonin,PCT)、C反应蛋白(C-reactive protein,CRP)和白细胞。
1.3 方法使用半自动微滴式数字PCR系统(D3200/7,杭州领航基因科技有限公司),包括:微滴生成仪DG32、扩增仪TC1、生物芯片阅读仪CS7。可以检测大肠埃希菌、铜绿假单胞菌、肺炎克雷伯菌、鲍曼不动杆菌、金黄色葡萄球菌、肠球菌属、链球菌属、嗜麦芽窄食单胞菌、阴沟肠杆菌、奇异变形杆菌、凝固酶阴性葡萄球菌、黏质沙雷菌、沙门菌属、柠檬酸杆菌属、脆弱拟杆菌、摩根摩根菌和洋葱伯克霍尔德菌等临床相关的17个细菌种/属,以及念珠菌属和blaKPC、mecA、blaNDM、blaIMP、blaOXA-48、vanA和vanM 7种耐药基因,涵盖了超过80%引发血流感染的致病菌[9]。在疑似血流感染患者使用抗生素之前,尽早抽取其10 mL静脉血到EDTA抗凝管(美国BD公司)内。随后3 000 r/min离心5 min,吸出2 mL血浆,加入3 mL裂解液、10 μL内参、30 μL磁珠和200 μL蛋白酶K,再经过各类缓冲液的漂洗和处理之后,使用60 μL洗脱液获得核酸,然后从中取出5 μL核酸,将其转移至含有10 μL扩增系统的反应管中,进行液滴制备和PCR扩增。扩增过程包括以下循环参数:37 ℃ 2 min;95 ℃ 5 min;然后95 ℃ 15 s、60 ℃ 30 s进行40个循环;最后25 ℃ 10 min。然后将芯片放入扫描仪中读取结果。对于血培养法,实验室将同步抽取的血培养瓶置于全自动血培养仪BACT/ALERT Vitruo 2(法国生物梅里埃公司)中进行培养,待仪器报告阳性结果后,将阳性瓶中的肉汤转种于血平板上,于35 ℃、体积分数5%二氧化碳条件下孵育24 h后,运用质谱仪VITEK MS(法国生物梅里埃公司)进行菌种鉴定,使用全自动微生物药敏仪VITEK 2(法国生物梅里埃公司)进行药敏试验。
结果解释:若检测到一种或多种病原体的基因拷贝数,则认为ddPCR结果为阳性,若没有检测到病原体则被认为是阴性。如果血培养和ddPCR的结果均为阳性或者阴性,则认为两者结果一致。当血培养结果为阳性、ddPCR结果为阴性时,则认为ddPCR的结果为假阴性;当血培养结果为阴性但ddPCR阳性时,结果归为可能的血流感染或者假阳性。
1.4 统计学分析使用GraphPad Prism 9软件进行统计学分析。正态分布的计量资料以均数±标准差(X±S)描述,偏态分布的计量资料以中位数(下4分位数Q25、上4分位数Q75)描述,采用卡方检验评估独立二项式变量,Mann-Whitney测试结果来比较ddPCR亚组之间的差异。P < 0.05表示差异有统计学意义。
2 结果 2.1 患者临床特征本研究在2021年12月—2023年8月于复旦大学附属闵行医院重症监护室收集55例疑似血流感染患者的血浆样本,同时进行血培养和ddPCR检测。入组患者的基线特征如表 1所示,男性37人,女性18人,平均年龄为73.27±15.06岁。患者的PCT、CRP、白细胞计数和中性粒细胞比例大部分高于正常参考范围(见表 1),占比分别为93.62%、93.75%、75.26%和92.47%。
| Clinical characteristics | Value | Reference ranges |
| Age (years) | 73.27±15.06 | N/A |
| Sex | ||
| Male[n(%)] | 37 (67.27%) | N/A |
| Female [n(%)] | 18 (32.73%) | N/A |
| Blood laboratory examination | ||
| PCT (ng/mL) | 17.18±28.76 | 0~0.065 |
| CRP (mg/L) | 108.15±70.04 | 0~10 |
| WBC (×109/L) | 14.92±8.18 | 3.5~9.5 |
| Neutrophil (%) | 87.47±7.74 | 40~75 |
在55例疑似血流感染的患者中(一个病人在治疗过程中可能多次同时进行血培养和ddPCR检测,所以共收集了血培养和ddPCR结果各100份),血培养阳性率为26%(26/100),ddPCR阳性率为70%(70/100)。其中,有23例样本血培养和ddPCR的结果同时阳性,不过其中有5例的病原体不一致(见表 2),27例为血培养和ddPCR的结果同时阴性,两者检测结果的符合率为45%(45/100)。ddPCR阳性而血培养阴性的标本有47例,其中32例还做了其他样本的培养,其结果与ddPCR的符合率为37.5%(12/32;见表 3);血培养阳性而ddPCR阴性的有3例。以血培养为诊断标准,ddPCR敏感性为69.2%(18/26),特异性为36.5%(27/74),阳性预测值(positive predictive value,PPV)为32.9%(23/70),阴性预测值(negative predictive value,NPV)为90%(27/30)。运用卡方检验得出ddPCR阳性率显著高于血培养(P < 0.01)。
| Sample ID | Blood culture | Droplet digital PCR(copies/mL) |
| 7 | Klebsiella pneumoniae | Klebsiella pneumoniae (156 564), Escherichia coli (4 811), Streptococcus (506) |
| 12 | Klebsiella pneumoniae | Klebsiella pneumoniae (32 090) |
| 26 | Enterococcus faecalis | Acinetobacter baumannii (1 393) |
| 28 | Candida glabrata | Staphylococcus aureus (71), Enterococcus (98), Candida (124) |
| 33 | Escherichia coli | Escherichia coli (802) |
| 45 | Citrobacter freudii | Citrobacter (5 578), Klebsiella (404 883), Candida (205) |
| 59 | Staphylococcus epidermidis, Staphylococcus capitis | Coagulase negative Staphylococcus (183) |
| 60 | Staphylococcus hemolytic, Enterococcus faecalis, Staphylococcus epidermidis, Enterococcus faecium | Coagulase negative Staphylococcus (61), Enterococcus (150) |
| 61 | Candida tropicalis | Candida (71 404), Coagulase negative Staphylococcus (44), Klebsiella (94) |
| 66 | Klebsiella pneumoniae | Klebsiella (3 174), Enterobacter cloacae (78), Coagulase negative (180), Candida (8 074) |
| 67 | Stenotrophomonas maltophilia | Stenotrophomonas maltophilia (9 090), Acinetobacter baumannii (4 869), Enterococcus (48) |
| 68 | Stenotrophomonas maltophilia | Stenotrophomonas maltophilia (31 214), Klebsiella (537) |
| 71 | Klebsiella pneumoniae | Klebsiella (123 921) |
| 73 | Proteus mirabilis | Proteus mirabilis (4 015), Streptococcus (209) |
| 74 | Staphylococcus capitis | Klebsiella (53), Acinetobacter baumannii (90) |
| 78 | Pseudomonas aeruginosa | Escherichia coli (58), Acinetobacter baumannii (58), Candida (40) |
| 83 | Candida glabrata | Candida (89), Escherichia coli (116), Enterococcus (550), streptococcus (790) |
| 84 | Bacteroides thetaiotaomicron | Bacteroides (1 336), Escherichia coli (45 000), Streptococcus (47) |
| 85 | Staphylococcus epidermidis | Streptococcus (58) |
| 86 | Klebsiella pneumoniae | Klebsiella (1 046), Escherichia coli (152), Streptococcus (46) |
| 91 | Staphylococcus hemolytic | Pseudomonas aeruginosa (5 524), Escherichia coli (100), Klebsiella (69) |
| 95 | Escherichia coli | Escherichia coli (202 612) |
| 100 | Escherichia coli | Escherichia coli (9 279), Enterococcus (186) |
| Sample ID | Droplet digital PCR (copies/mL) |
Blood culture | Culture of other samples |
| 1 | Enterococcus (117.9) | Negative | Acinetobacter baumannii |
| 2 | Coagulase negative Staphylococcus (174.6), Streptococcus (174.6), Candida (87.3) | Negative | Acinetobacter baumannii |
| 4 | Coagulase negative Staphylococcus (119), Streptococcus (285) | Negative | Acinetobacter baumannii |
| 8 | Klebsiella pneumoniae (30 600), Escherichia coli (907) | Negative | Negative |
| 9 | Acinetobacter baumannii (1 393) | Negative | Negative |
| 10 | Klebsiella pneumoniae (2 730), Escherichia coli (133), Enterococcus (634) | Negative | No other culture |
| 11 | Klebsiella (2 642), Streptococcus (803) | Negative | No other culture |
| 13 | Klebsiella (1 142), Streptococcus (756), Enterococcus (149) | Negative | No other culture |
| 14 | Klebsiella (965), Streptococcus (401) | Negative | Candida albicans |
| 15 | Klebsiella (170), Enterococcus (130) | Negative | No other culture |
| 16 | Klebsiella (1 418), Escherichia coli (76), Acinetobacter baumannii (1 375) | Negative | No other culture |
| 17 | Klebsiella (2 445) | Negative | No other culture |
| 18 | Klebsiella (193) | Negative | No other culture |
| 19 | Klebsiella (515) | Negative | Klebsiella pneumoniae |
| 20 | Enterococcus (88) | Negative | No other culture |
| 21 | Acinetobacter (40), Streptococcus (86) | Negative | Klebsiella pneumoniae |
| 22 | Acinetobacter (1 171), Klebsiella (347) | Negative | No other culture |
| 23 | Coagulase negative Staphylococcus (95), Streptococcus (113) | Negative | Pseudomonas aeruginosa, Proteus mirabilis |
| 35 | Escherichia coli (112) | Negative | No other culture |
| 39 | Enterobacter cloacae (109.8), Klebsiella (116), Escherichia coli (287.1), Citrobacter (143.1) | Negative | Klebsiella pneumoniae |
| 44 | Coagulase negative Staphylococcus (50) | Negative | Negative |
| 46 | Klebsiella (19 616), Acinetobacter baumannii (175) | Negative | Klebsiella pneumoniae |
| 47 | Klebsiella (1 527) | Negative | No other culture |
| 48 | Klebsiella (1 386) | Negative | No other culture |
| 51 | Escherichia coli (144.9), Streptococcus (62.1) | Negative | Acinetobacter baumannii |
| 53 | Acinetobacter baumannii (680), Candida (80) | Negative | Acinetobacter baumannii |
| 54 | Acinetobacter baumannii (337.5), Enterococcus (101.7) | Negative | No other culture |
| 56 | Klebsiella (70), Enterococcus (53) | Negative | Klebsiella pneumoniae |
| 57 | Klebsiella (136.8), Enterococcus (90), Coagulase negative Staphylococcus (291.6), Streptococcus (190.8) | Negative | Klebsiella pneumoniae |
| 62 | Acinetobacter baumannii (44), Coagulase negative Staphylococcus (120), Enterococcus (139), Streptococcus (102) | Negative | Acinetobacter baumannii |
| 63 | Escherichia coli (61), Acinetobacter baumannii (1 242), Enterococcus (48), Streptococcus (202), Candida (87) | Negative | Acinetobacter baumannii, Enterococcus faecium |
| 64 | Enterococcus (180) | Negative | Enterococcus faecium |
| 69 | Coagulase negative Staphylococcus (27.45) | Negative | Acinetobacter baumannii |
| 70 | Acinetobacter baumannii (26.55) | Negative | Negative |
| 72 | Klebsiella (14 670) | Negative | Klebsiella pneumoniae |
| 75 | Klebsiella (50), Enterococcus (47), Candida (52) | Negative | Negative |
| 79 | Klebsiella (68), Enterococcus (152) | Negative | Klebsiella pneumoniae |
| 80 | Escherichia coli (44), Enterococcus (1 270) | Negative | Pseudomonas aeruginosa |
| 82 | Escherichia coli (496), Streptococcus (3 356) | Negative | Negative |
| 87 | Escherichia coli (900), Streptococcus (45) | Negative | Negative |
| 88 | Candida (50), Streptococcus (60) | Negative | Negative |
| 89 | Streptococcus (69) | Negative | Negative |
| 90 | Enterococcus (60) | Negative | No other culture |
| 93 | Staphylococcus aureus (3 600) | Negative | Negative |
| 94 | Acinetobacter baumannii (50), Streptococcus (45) | Negative | Negative |
| 96 | Klebsiella (128.5), Escherichia coli (64), Acinetobacter baumannii (27.5), Serratia marcescens (53) | Negative | Negative |
| 97 | Enterococcus (81) | Negative | No other culture |
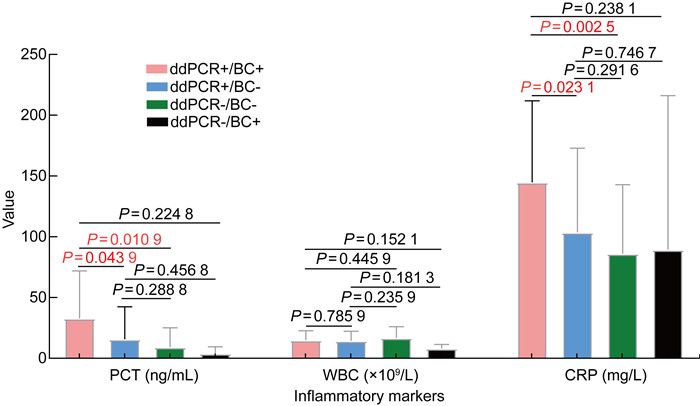
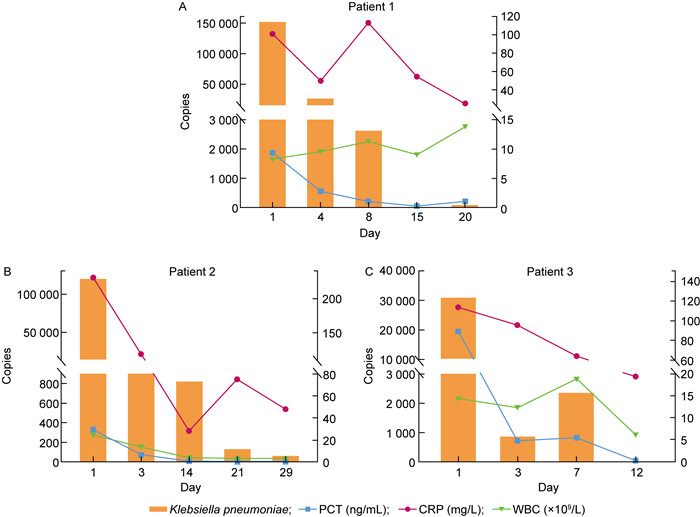
本研究比较分析了ddPCR+/BC+、ddPCR+/ BC-、ddPCR-/BC+、ddPCR-/BC- 组的炎症指标。结果显示:ddPCR+/BC+组的PCT、CRP显著高于ddPCR+/BC-和ddPCR-/BC-组(P=0.043 9,P=0.010 9;P=0.023 1,P=0.002 5),但和ddPCR-/BC+组间没有显著性差异(P=0.224 8,P=0.238 1);表明ddPCR+/BC+组和ddPCR-/BC+组血清中的炎性特征相近。ddPCR+/BC+组的白细胞和其他3组亦无显著性差异(P=0.785 9,P=0.152 1,P=0.445 9),表明在4组数据中,白细胞的数值与血流感染的诊断没有明显的相关性。ddPCR+/BC-组的PCT、CRP、白细胞和ddPCR-/BC-组无显著性差异(P=0.288 8,P=0.291 6,P=0.235 9),表明ddPCR+/BC-组和ddPCR-/BC-组血清中的炎性特征相近(见图 1)。同时本文对ddPCR+/BC+组中的3个病例进行了4~5个不同病程时间点的采样,并使用ddPCR进行动态监测,结果发现ddPCR的基因拷贝数与PCT、CRP和白细胞的指标值变化趋势相似,即当患者接受有效治疗后,ddPCR的基因拷贝数和PCT、CRP和白细胞基本呈同步下降的趋势(见图 2)。
|
| ddPCR: droplet digital PCR. BC: blood culture. 图 1 ddPCR+/BC+、ddPCR+/BC-、ddPCR-/BC-和ddPCR-/BC+组的炎性标志物比较 Fig. 1 Comparison of the inflammatory markers in groups of ddPCR+/BC+, ddPCR+/BC-, ddPCR-/BC-, and ddPCR-/BC+ |
|
| 图 2 3个病例中ddPCR基因载量与降钙素原、C反应蛋白和白细胞的变化趋势 Fig. 2 The trends of gene loads in ddPCR test changing with procalcitonin (PCT), C-reactive protein (CRP), and white blood cells (WBC) values in three patients |
本研究中,ddPCR共检出18份标本含有耐药基因,其中11份为mecA基因(与甲氧西林耐药相关),5份为blaKPC,1份为blaNDM和blaIMP,1份同时携带blaKPC、blaNDM和blaIMP基因。与ddPCR结果相比,血培养阳性菌株中共检出3株甲氧西林耐药的葡萄球菌(3 vs 11)和6株碳青霉烯耐药的肺炎克雷伯菌(6 vs 7)(见表 4),数量都较少;有9份标本ddPCR检出耐药基因而血培养未检出耐药菌(见表 5)。以上结果提示:对于耐药基因的检测,ddPCR比常规药敏试验具有更高的灵敏度,可作为血培养的重要补充。
| Antibiotics resistance gene detected by droplet digital PCR | Pathogens | Antibiotics resistant bacteria cultured from blood and other cultures |
| blaKPC (n=5) | Klebsiella pneumoniae | CRKP (n=4) |
| blaNDMand blaIMP (n=1) | Klebsiella pneumoniae | CRKP (n=1) |
| blaKPC, blaNDM and blaIMP (n=1) | Klebsiella pneumoniae | CRKP (n=1) |
| mecA (n=11) | MRS | MRS (n=3) |
| MRS:methicillin resistant Staphylococcus;CRKP:carbapenem-resistant Klebsiella pneumonia. | ||
| Sample ID | Droplet digital PCR (copies/mL) | Blood culture |
| 4 | Coagulase negative Staphylococcus (119), Streptococcus (285), mecA (107) | Negative |
| 8 | Klebsiella pneumoniae (30 600), Escherichia coli (907), mecA (95) | Negative |
| 9 | Acinetobacter baumannii (1 393), mecA (183) | Negative |
| 16 | Klebsiella pneumoniae (1 418), Escherichia coli (76), Acinetobacter baumannii (1 375), mecA (240) | Negative |
| 26 | Acinetobacter baumannii (1 393), mecA (76) | Enterococcus faecalis |
| 28 | Staphylococcus aureus (71), Enterococcus (98), Candida (124), mecA (158) | Candida glabrata |
| 57 | Klebsiella (136.8), Enterococcus (90), Coagulase negative Staphylococcus (291.6), Streptococcus (190.8), mecA (639), blaKPC (98.4), blaNDM (74.7), blaIMP (74.7) | Negative |
| 62 | Acinetobacter baumannii (44), Coagulase negative Staphylococcus (120), Enterococcus (139), Streptococcus (102), mecA (114) | Negative |
| 10 | Klebsiella pneumoniae (2 730), Escherichia coli (133), Enterococcus (634), blaKPC (75) | Negative |
经验性运用抗生素治疗、血液中细菌载量本身偏低,导致血培养阳性率低和检出周期较长,从而无法及时诊断出血流感染的患者,这均会影响患者的治疗和预后[10]。随着分子技术的不断发展,快速诊断病原体的方法亦越来越多。基质辅助激光解吸电离飞行时间质谱(matrix-assisted laser desorption ionization time-of-flight mass spectrometry,MALDI-TOF MS)具有高灵敏度和特异性,但其准确性可能会受到数据库的影响[11]。宏基因组二代测序(metagenomics next-generation sequencing,mNGS)结果的阳性率比血培养高6倍左右,特别是对罕见细菌和难以培养得到的病原体,但其检测费用高昂,并且需要专业的生物信息学分析团队来处理和解释测序数据,这些因素导致周转时间较长,mNGS在临床实践中存在一定缺陷[12]。
ddPCR是一种新的分子诊断技术,相较于传统的PCR,可以做到病原体的及时定量检出,并实现动态监测,将病原诊断时间缩短至3 h左右。它已被广泛应用于肿瘤检测和产前诊断,并已在感染性疾病领域有所尝试[13-15]。ddPCR具有灵敏度高、特异性强和采血量少的优点,尤其适用于接受经验性抗生素治疗的患者。
在ddPCR和血培养的结果同时阳性时,ddPCR显示多种微生物感染的发生率较高,达到69.57%,阳性率高于血培养(8.70%;P < 0.01)(见表 2)。究其原因,可能由于ddPCR灵敏度高,病原体浓度达到10 CFU/mL即可检出,患者使用抗生素之后病原体仍可被检出[8]。而血培养仅能检测出生长最快的细菌,而量少、生长缓慢的细菌由于营养被掠夺而无法检出[16]。以血培养为诊断血流感染的金标准,ddPCR的灵敏度为88.5%,阴性预测值达90%;来自浙江的研究显示,ddPCR的灵敏度和阴性预测值更高,达到100%[17]。然而,由于ddPCR仅能检测出基因片段,而无法区分该片段来自活体微生物还是凋亡微生物,故存在一定的假阳性。另一方面,ddPCR+/BC-组的其他标本的培养结果与ddPCR的结果符合率接近40%(见表 3),提示ddPCR存在检出其他部位病原体释放入血的游离DNA的可能。ddPCR+/BC+组的白细胞和其他3组相比没有显著性差异(见图 1),提示在本研究中,白细胞和血流感染的检出率很可能没有相关性。ddPCR+/BC+组的PCT、CRP显著高于ddPCR+/BC-和ddPCR-/BC-组,而和ddPCR-/BC+组间没有显著性差异(见图 1),表明PCT和CRP与血培养阳性结果有显著相关性,而与ddPCR阳性结果没有相关性,这提示血培养检测阴性的非严重的血流感染患者仍可能受益于ddPCR检测,从而获得更为及时的治疗[17]。
分析连续采样的3个病例的数据,可知ddPCR的基因拷贝数与PCT和CRP的值变化趋势相似(见图 2),即当患者接受有效治疗后,ddPCR的基因拷贝数和PCT、CRP同步下降。该现象也在华山医院和南京鼓楼医院的研究中得到证实[8-9]。由此看来,ddPCR的定量检出结果可以作为血流感染患者病原体清除的依据,临床可被用作患者预后及抗生素更换的参考。
病原体对抗菌药物的耐药性会导致血流感染患者治疗的失败,既有研究通过筛选耐药基因,认为blaKPC、blaNDM和blaOXA-48是影响治疗革兰氏阴性菌血流感染的关键因素,而mecA和vanA是影响治疗革兰氏阳性菌血流感染的关键因素[18]。所以,早期快速诊断出耐药基因以指导抗生素用药,对减轻血流感染患者的症状至关重要[19]。本研究共检出18份标本携带耐药基因,与甲氧西林和碳青霉烯类的耐药性相关,检出率高于血培养和其他标本培养物的检出率(18 vs 9),而传统培养方法的低检出率可能是由标本中细菌含量低所致。ddPCR在检测耐药基因方面灵敏度高,以blaKPC基因为例,低至80 copies/mL即可被检出[20]。尽管如此,由于细菌耐药的机制非常复杂,有些耐药基因在不同菌株中的表达情况不同,导致基因型和耐药表型也存在不一致的情况,因此细菌药敏试验仍然是临床用药的重要依据[21]。目前来看,虽然ddPCR无法取代传统的血培养和药敏试验,但仍然是一种有潜力的诊断方法,可作为传统方法的重要补充。尤其在重症血流感染的患者中,可以发挥它快速、精确的优势[22]。
本研究具有一定的局限性。首先,样本量不够多,代表性略显不足,可能会导致研究结果出现一定的偏差。其次,由于微生物游离DNA可能是从死亡病原体中释放出来的,ddPCR检测到血液中微生物游离DNA,并不一定表明血液中存在活的病原体。再则,血培养检测的致病病原体覆盖范围更广,而ddPCR识别的目标病原体相对较少,因此,仅根据ddPCR的检测结果无法确定患者是否患有单一微生物感染或多种微生物感染。此外,区分致病病原体与正常微生物或环境污染物仍具有一定的挑战性。尽管ddPCR检测可以报告定量结果,但目前还没有明确的界限可区分感染与定植或污染物。因此,为了克服上述一部分限制,可通过扩大检测组以覆盖其他病原体并跟踪微生物游离DNA的载量变化,从而通过动态监测微生物的游离DNA浓度来反映血流感染的进展。在明确通过游离DNA测序检测到的病原体的临床意义时,应考虑患者整体的临床情况。
本研究表明,液滴数字PCR可以快速、灵敏和定量地检测疑似血流感染患者体中常见的病原体,能够在病原菌早期预警、疾病动态监测及抗生素运用指导方面发挥潜力。血培养依然是血流感染诊断的金标准,ddPCR可以作为血培养检测的重要补充,为重症疑似血流感染患者做出快速、准确的诊断,同时耐药基因的检测能够为临床用药提供有价值的参考。
| [1] |
Leibovici-Weissman Y, Tau N, Yahav D. Bloodstream infections in the elderly: what is the real goal?[J]. Aging Clin ExpRes, 2021, 33(4): 1101-1112.
[DOI]
|
| [2] |
Seok H, Song J, Jeon JH, Choi HK, Choi WS, Moon S, Park DW. Timing of antibiotics in septic patients: a prospective cohort study[J]. Clin Microbiol Infect, 2020, 26(11): 1495-1500.
[DOI]
|
| [3] |
Ferrer R, Martin-Loeches I, Phillips G, Osborn TM, Townsend S, Dellinger RP, Artigas A, Schorr C, Levy MM. Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour: results from a guideline-based performance improvement program[J]. Crit Care Med, 2014, 42(8): 1749-1755.
[DOI]
|
| [4] |
Della Rocca MT, Panetta V, Durante A, Bucci L, Matano A, Annecchiarico A, Greco R. Pathogens distribution and antimicrobial resistance pattern of blood stream infections in Southern Italian hospital, 2016-2021 surveillance[J]. New Microbiol, 2023, 46(1): 29-36.
[URI]
|
| [5] |
Li Y, Ma M, Xu X, Li Q, Ji C. Value of digital PCR in the early diagnosis of sepsis: a systematic review and meta-analysis[J]. Crit Care, 2022, 72: 154138.
[DOI]
|
| [6] |
Song Y, Neff M, Gyarmati P. Challenges and advances in the diagnosis of bloodstream infection[J]. Future Microbiol, 2022, 17: 311-314.
[DOI]
|
| [7] |
Li H, Bai R, Zhao Z, Tao L, Ma M, Ji Z, Jian M, Ding Z, Dai X, Bao F, Liu A. Application of droplet digital PCR to detect the pathogens of infectious diseases[J]. Biosci Rep, 2018, 38(6): BSR20181170.
[DOI]
|
| [8] |
Lin K, Zhao Y, Xu B, Yu S, Fu Z, Zhang Y, Wang H, Song J, Fan M, Zhou Y, Ai J, Qiu C, Zhang H, Zhang W. Clinical diagnostic performance of droplet digital PCR for suspected bloodstream infections[J]. Microbiol Spectr, 2023, 11(1): e0137822.
[DOI]
|
| [9] |
Li M, Zhao L, Zhu Y, Ou M, Xu H, Hu X, Wei H, Chen Y, Shen H. Clinical value of droplet digital PCR in the diagnosis and dynamic monitoring of suspected bacterial bloodstream infections[J]. Clin Chim Acta, 2023, 550: 117566.
[DOI]
|
| [10] |
Zornitzki L, Anuk L, Frydman S, Morag-Koren N, Zahler D, Freund O, Biran R, Liron Y, Tau L, Tchebiner JZ, Katash H, Bornstein G. Rate and predictors of blood culture positivity after antibiotic administration: a prospective single-center study[J]. Infection, 2024, 52(2): 483-490.
[DOI]
|
| [11] |
Tsuchida S, Umemura H, Nakayama T. Current status of matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS) in clinical diagnostic microbiology[J]. Molecules, 2020, 25(20): 4775.
[DOI]
|
| [12] |
Rodino KG, Simner PJ. Status check: next-generation sequencing for infectious-disease diagnostics[J]. J Clin Invest, 2024, 134(4): e178003.
[DOI]
|
| [13] |
Olmedillas-López S, Olivera-Salazar R, García-Arranz M, García-Olmo D. Current and emerging applications of droplet digital PCR in oncology: an updated review[J]. Mol Diagn Ther, 2022, 26(1): 61-87.
[DOI]
|
| [14] |
Sawakwongpra K, Tangmansakulchai K, Ngonsawan W, Promwan S, Chanchamroen S, Quangkananurug W, Sriswasdi S, Jantarasaengaram S, Ponnikorn S. Droplet-based digital PCR for non-invasive prenatal genetic diagnosis of α and β-thalassemia[J]. Biomed Rep, 2021, 15(4): 82.
[DOI]
|
| [15] |
Chen B, Jiang Y, Cao X, Liu C, Zhang N, Shi D. Droplet digital PCR as an emerging tool in detecting pathogens nucleic acids in infectious diseases[J]. Clin Chim Acta, 2021, 517: 156-161.
[DOI]
|
| [16] |
Hu B, Tao Y, Shao Z, Zheng Y, Zhang R, Yang X, Liu J, Li X, Sun R. A comparison of blood pathogen detection among droplet digital PCR, metagenomic next-generation sequencing, and blood culture in critically Ill patients with suspected bloodstream infections[J]. Front Microbiol, 2021, 12: 641202.
[DOI]
|
| [17] |
Zheng Y, Jin J, Shao Z, Liu J, Zhang R, Sun R, Hu B. Development and clinical validation of a droplet digital PCR assay for detecting Acinetobacter baumannii and Klebsiella pneumoniae in patients with suspected bloodstream infections[J]. Microbiologyopen, 2021, 10(6): e1247.
[DOI]
|
| [18] |
Russotto V, Cortegiani A, Graziano G, Saporito L, Raineri SM, Mammina C, Giarratano A. Bloodstream infections in intensive care unit patients: distribution and antibiotic resistance of bacteria[J]. Infect Drug Resist, 2015, 8: 287-296.
[DOI]
|
| [19] |
Carlesse F, Cappellano P, Quiles MG, Menezes LC, Petrilli AS, Pignatari AC. Clinical relevance of molecular identification of microorganisms and detection of antimicrobial resistance genes in bloodstream infections of paediatric cancer patients[J]. BMC Infect Dis, 2016, 16(1): 462.
[DOI]
|
| [20] |
Wu J, Tang B, Qiu Y, Tan R, Liu J, Xia J, Zhang J, Huang J, Qu J, Sun J, Wang X, Qu H. Clinical validation of a multiplex droplet digital PCR for diagnosing suspected bloodstream infections in ICU practice: a promising diagnostic tool[J]. Crit Care, 2022, 26(1): 243.
[DOI]
|
| [21] |
Wenzler E, Maximos M, Asempa TE, Biehle L, Schuetz AN, Hirsch EB. Antimicrobial susceptibility testing: an updated primer for clinicians in the era of antimicrobial resistance: insights from the society of infectious diseases pharmacists[J]. Pharmacotherapy, 2023, 43(4): 264-278.
[DOI]
|
| [22] |
Lin K, Zhang HC, Zhao YH, Xia J, Ai JW, Zhang WH. The direct application of plasma droplet digital PCR in the ultra-early pathogen detection and warning during sepsis: case reports[J]. J Infect Public Health, 2022, 15(4): 450-454.
[DOI]
|
 2024, Vol. 19
2024, Vol. 19


